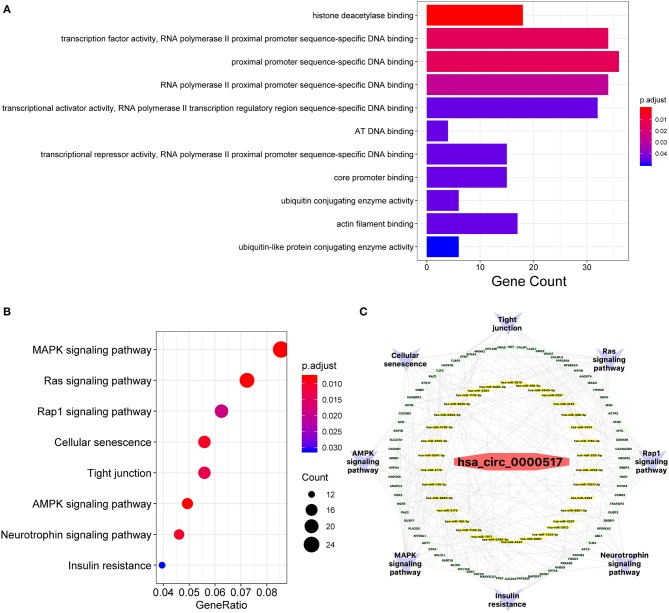
Figure 4.
GO and KEGG functional analyses. (A) Gene ontology (GO) analysis of potential targets of miRNAs. The biological process (BP), cellular component (CC), and molecular function (MF) of potential targets were clustered based on ClusterProfiler package in R software (version: 3.12.0). The abscissa strands for gene counts and the ordinate means the enriched pathways. (B) The enriched KEGG signaling pathways were selected to demonstrate the primary biological actions of major potential targets. The abscissa indicates gene ratio and the enriched pathways were presented in the ordinate. (C) The has_circ_0000517 regulatory Network. The pathways, hub genes, miRNAs were presented in outer circle, middle circle and inner circle, respectively.