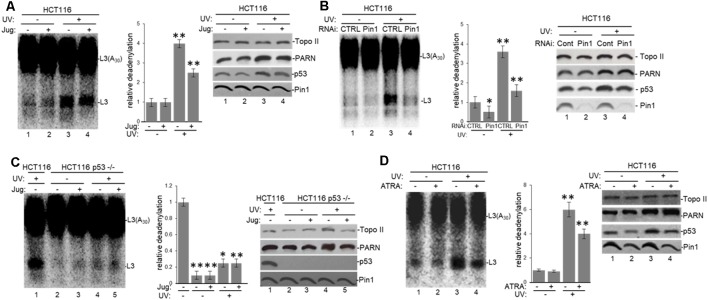
Figure 6.
Pin1 is part of a deadenylation activator complex that includes PARN, tau and p53 under damaging conditions. (A,B) Levels of nuclear deadenylation correlate with Pin1 activity and expression in HCT116 cells. NEs from cells incubated with either (A) 5 μM juglone for 2 h or (B) Pin1/control siRNAs for 48 h were used in deadenylation assays. Indicated samples were also treated with UV (40 Jm−2) and allowed to recover for 2 h. The deadenylation reactions were incubated for 90 min. RNAs were purified and analyzed by denaturing PAGE. Representative deadenylation reactions from three independent biological assays are shown. Positions of the polyadenylated RNA L3(A30) and the L3 deadenylated product are indicated. On the right, relative deadenylation (RD) levels, calculated as {L3 fragment/[L3 fragment + L3(A30)]} ×100, were plotted for each condition. The RD levels for control conditions were arbitrarily set at 1.0. The means ± standard deviation of RD values are indicated. The P-values are indicated as *(≤0.05) or **(≤0.005). Quantifications were done with ImageJ software (http://rsb.info.nih.gov/ij/). Right panels show Western blot analysis of indicated proteins from NEs tested for deadenylation. (C) Pin1-mediated activation of nuclear deadenylation is dependent on p53 expression. NEs from HCT116 or HCT116 p53−/− cells incubated with 5 μM juglone and/or treated with UV (40 Jm−2) and allowed to recover for 2 h were used in deadenylation assays. Deadenylation reactions were performed as described in (A,B). RD levels in samples from HCT116 cells were arbitrarily set at 1.0. Right panels show Western blot analysis of indicated proteins from NEs used in deadenylation assays. (D) Levels of nuclear deadenylation were affected by Pin1-inhibitor all-trans retinoic acid (ATRA) in HCT116 cells. NEs from cells incubated with 1 μM ATRA for 72 h were used in deadenylation assays. Indicated samples were also treated with UV (40 Jm−2) and allowed to recover for 2 h. Deadenylation reactions were performed as described in (A). RD level for untreated HCT116 cells was arbitrarily set at 1.0. Right panels show Western blot analysis of indicated proteins from NEs used in deadenylation assays.