Figure 1.
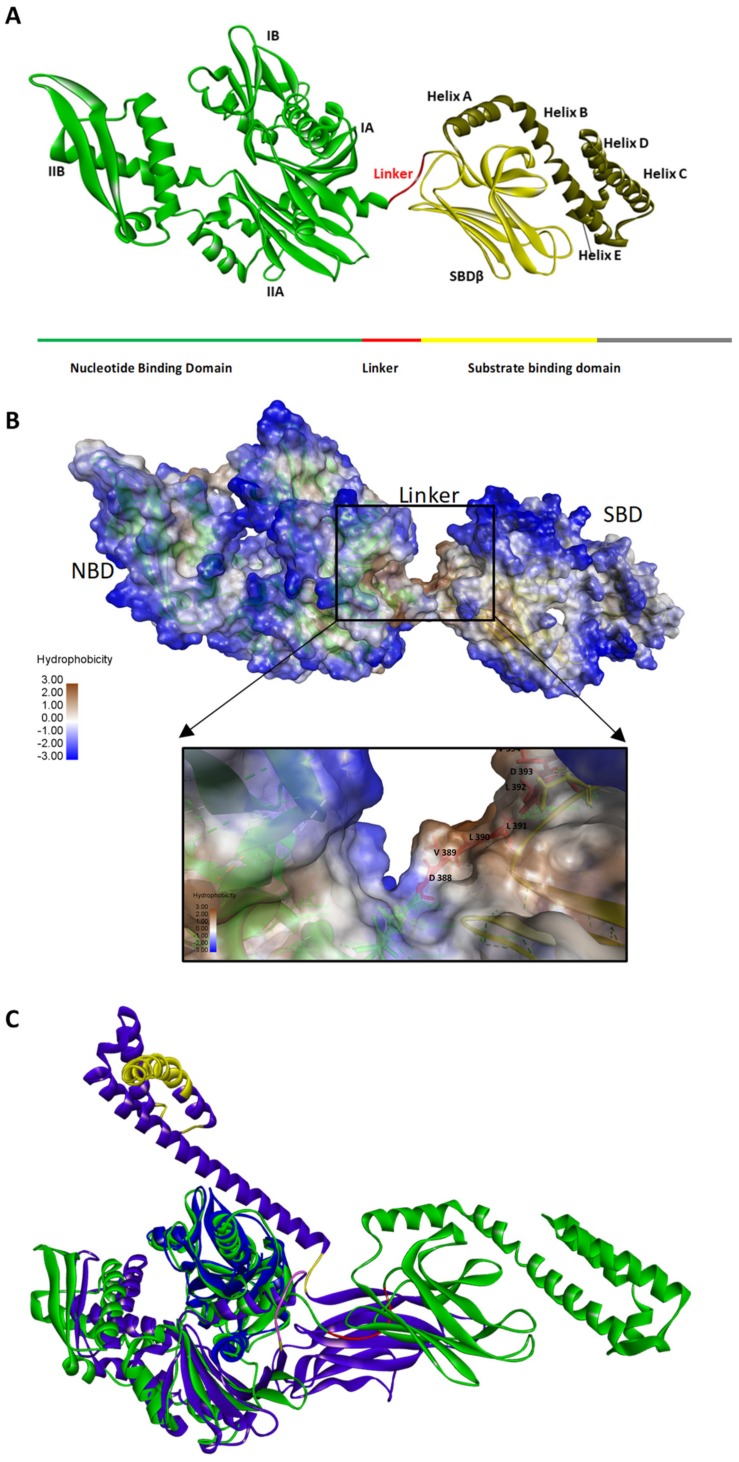
The domain organization of heat shock 70 (Hsp70). Three-dimensional model of E. coli Hsp70 (DnaK) showing domain organization (A). The N-terminal nucleotide binding domain (NBD) is made up of lobes IA, IIA, IB and IIB (Green). The substrate binding domain (SBD) is made of the SBDβ (yellow) which forms the substrate binding cleft and the SBDα which is characterized by helices A, B, C, D and E (brown). The linker connects the NBD and SBD of Hsp70s (red). The linker of canonical Hsp70s is typically hydrophobic in nature, as confirmed by after hydrophobicity analysis of residues using discovery studio visualizer (https://www.3dsbiovia.com). (B) A three-dimensional model of a canonical Hsp70 (green) with the linker highlighted (red). The three-dimensional model of a canonical Hsp70 (P0A648; green) was superimposed against that of Hsp110 (P32589; blue) whose linker (purple) and acidic insertions (yellow) are highlighted (C). The modelling was conducted using template C3c7n.1A.pdb [14] on Chimera [15].