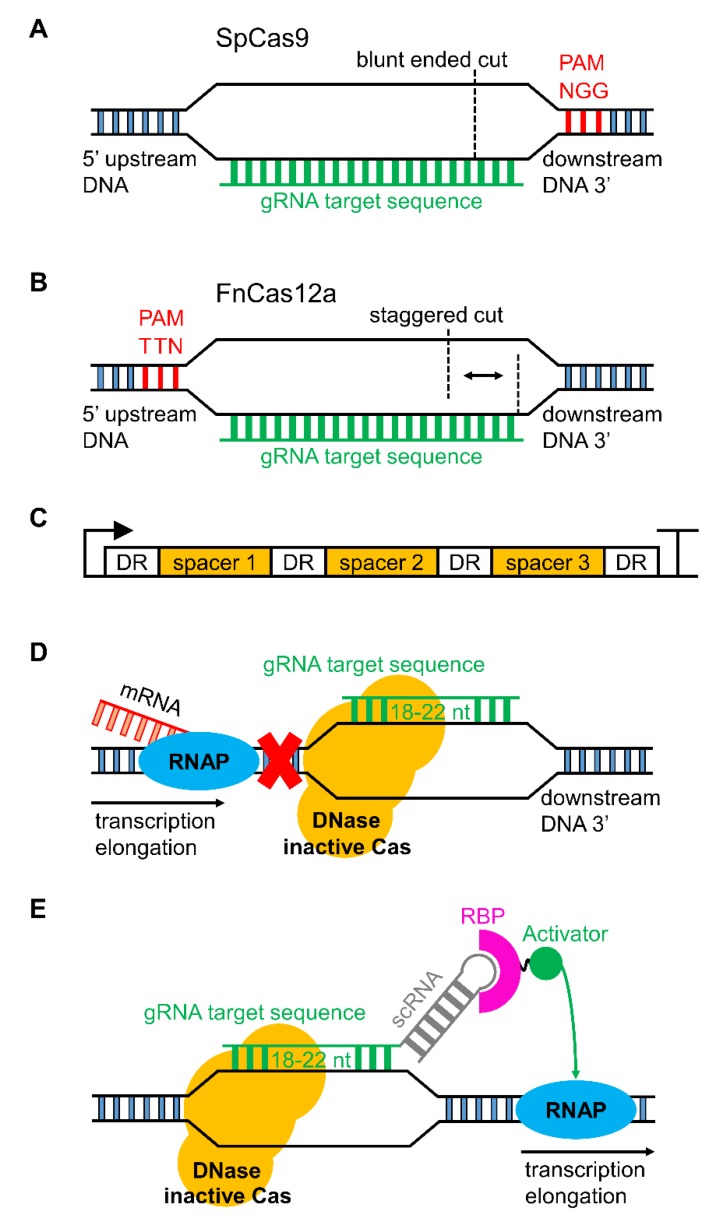
Figure 1.
Basic overview of CRISPR/Cas-based genome editing and CRISPR interference in cyanobacteria. (A) The type II-A CRISPR/Cas system comprising SpCas9 that is targeted by a guide RNA (gRNA, green) upstream of an NGG protospacer adjacent motif (PAM) (red). SpCas9 generates a blunt-ended double-stranded break (DSB) in DNA (black dashed line). (B) The type II-V CRISPR/Cas system comprising FnCas12a that is targeted by a gRNA downstream of a TTN PAM. Cas12a confers a staggered DSB in DNA that results in a five-nucleotide overhang. (C) Illustration of a single expression cassette containing three spacers (i.e., a spacer array, orange) that are flanked by direct repeat regions (DRs, white). Cas12a recognises and cleaves spacers in response to a DR to generate a mature gRNA from each spacer. (D) Overview of transcriptional inhibition by CRISPRi. DNase inactive Cas enzymes (e.g., dCas9 or ddCas12a) are targeted to a DNA locus by a gRNA (typical length indicated as 18–22 nucleotides), which blocks RNA polymerase (RNAP, blue) to prevent mRNA (red) synthesis. (E) Example of transcriptional activation by CRISPRa. DNase inactive Cas enzymes (e.g., dCas9 or ddCas12a) are targeted by a gRNA fused to a scaffold RNA (scRNA, grey). The scRNA recruits an RNA binding protein (RBP, pink) fused to a transcriptional activator (green) (e.g., [108]), which subsequently leads to activation of RNAP and transcription elongation.