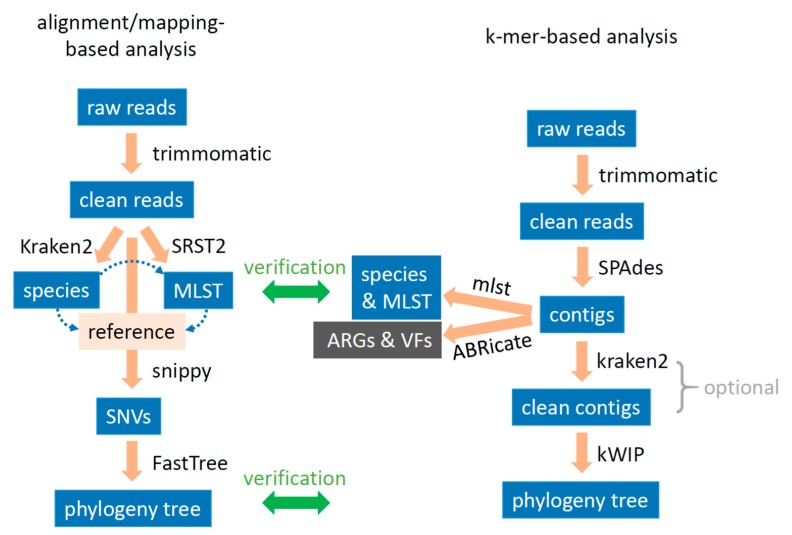
Figure 1.
Schematic bioinformatics data processing workflows for whole-genome sequence analysis. Left: alignment/mapping-based analysis. Multi-locus sequence typing (MLST) is based on species determination, while the reference genome is selected based on both species and MLST uncovered. Right: k-mer-based analysis. Contigs cleaning is optional, mainly for those isolates with an outstanding size of the assembled “genome”. SNVs: single nucleotide variants; ARGs: antibiotic resistance genes; VFs: virulence factors.