Figure 7.
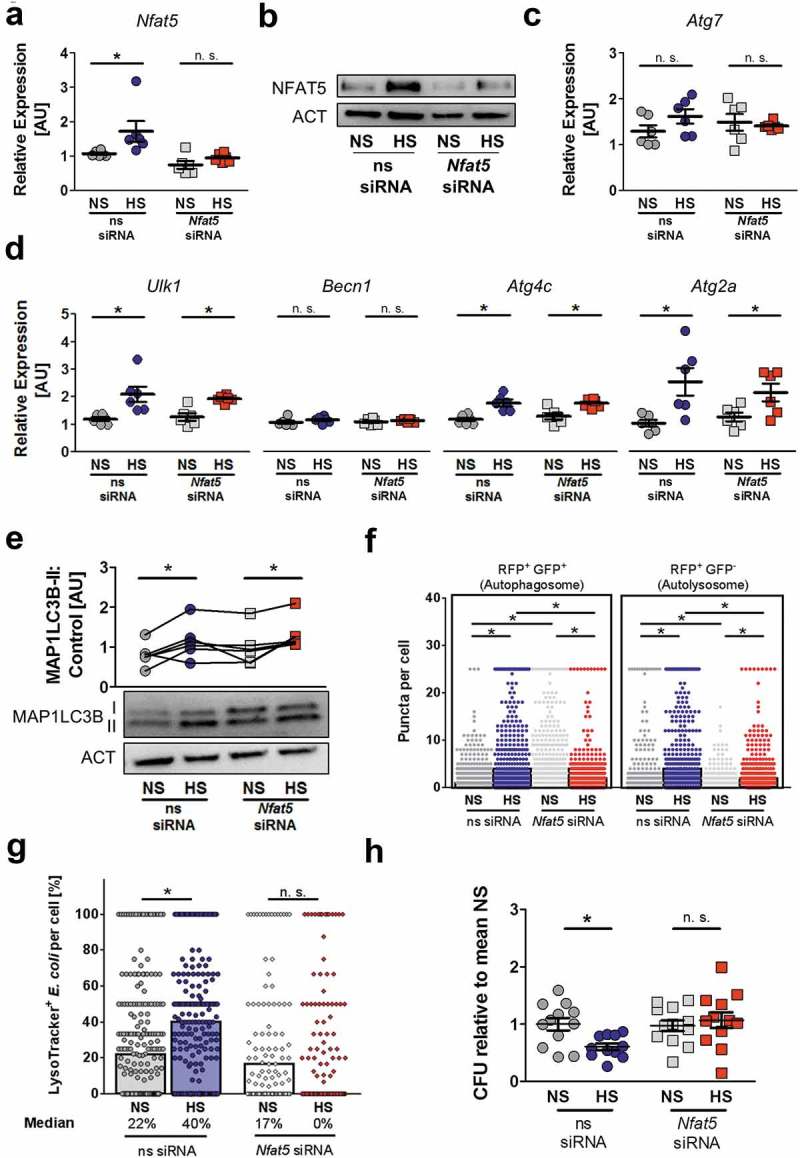
NFAT5 is required for HS-facilitated targeting of intracellular E. coli to autolysosomes. (a to d) As in Figure 1(f), but non-silencing siRNA (ns siRNA) and Nfat5-specific siRNA (Nfat5 siRNA)-treated RAW264.7 MΦ were used. (a) Nfat5 mRNA levels (means ± s.e.m; n = 6; Student’s t test or Mann-Whitney test, * p < 0.05). (b) Immunoblotting of NFAT5 and ACT. (c) As (a), but Atg7 mRNA levels (means ± s.e.m; n = 6; Student’s t test ± Welch correction). (d) As (a), but Ulk1, Becn1, Atg4c and Atg2a mRNA levels (means ± s.e.m; n = 6; Student’s t test ± Welch correction; * p < 0.05). (e) Immunoblotting and densitometry of MAP1LC3B/LC3 and ACT ½ h after infection (n = 6; paired Student’s t test or Wilcoxon signed rank test; *p < 0.05). (f) As in Figure 1(f), but ns siRNA- and Nfat5 siRNA-treated RFP-GFP-mLC3 RAW264.7 MΦ were used. RFP+ and RFP+GFP+-puncta were counted per cell. At least 290 cells from two independent experiments (bars: median scores; Kruskal Wallis test with Dunn’s Multiple Comparison Test, * p < 0.05). (g) As Figure 1(f), but RAW264.7 MΦ were used and stained with LysoTracker. Colocalization of sfGFP-E. coli and LysoTracker per cell. At least 101 cells from at least two independent experiments (bars: median scores; Kruskal Wallis test with Dunn’s Multiple Comparison Test, * p < 0.05). (h) As in Figure 1(a), but ns siRNA- and Nfat5 siRNA-treated RAW264.7 MΦ were used. Intracellular E. coli load in CFU relative to mean CFU under NS conditions (means ± s.e.m; n = 11–12; Student’s t test ± Welch correction, * p < 0.05).
