Figure 1.
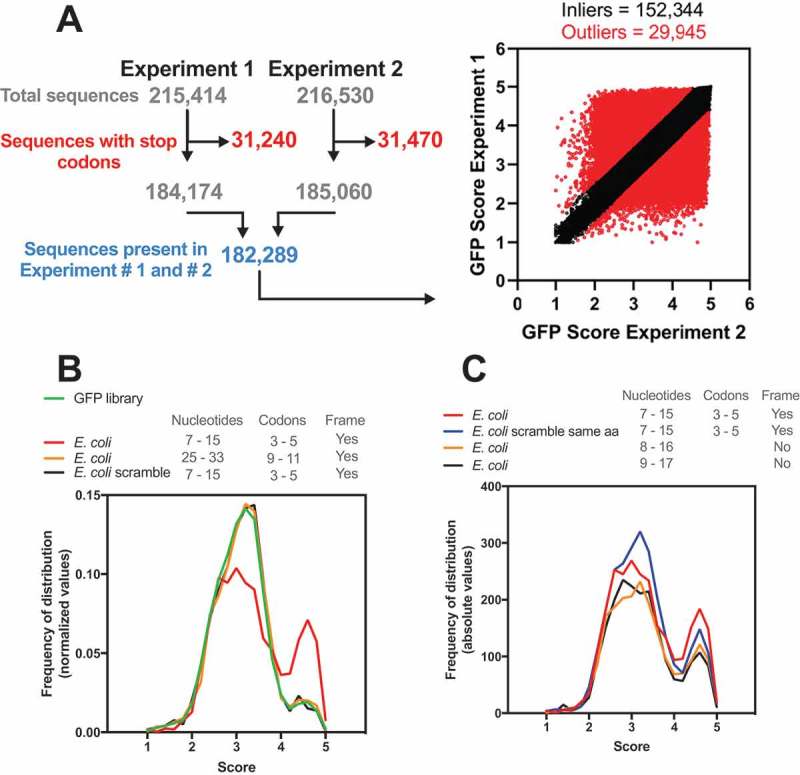
The E. coli genome has a bias towards nucleotide composition at codons 3–5. (A) Djuranovic’s group performed a high-throughput screen varying the nucleotide composition at codons 3–5 in GFP. In experiments #1 and #2, 215,414 and 261,530 different compositions were analysed, respectively, regarding the GFP fluorescence levels. The sequences with a stop codon (31,240 and 31,470 for experiments #1 and #2, respectively) were removed, and only sequences present in both experiments were used (182,289). The outliers (29,945) were defined by setting Q = 1% in the linear regression. We then calculated the average GFP score of the inliers (152,344) from experiments #1 and #2. This list was used in all subsequent bioinformatics experiments. (B) Density histogram of GFP scores for genes identified in E. coli. The nucleotide composition at codon positions 3–5 or 9–11 was analysed. As a control, we used a scrambled genome where the codon proportion was maintained, but their position was randomly changed. Note that only codon positions 3–5 in the real genome possessed a bias towards high GFP scores. (C) The effect of amino acid composition and mRNA sequence on GFP score bias was analysed. As a control, we used a scrambled genome where the codons were randomly changed, keeping the codon proportion and amino acid sequence of each gene (E. coli scramble same aa).
