Figure 3.
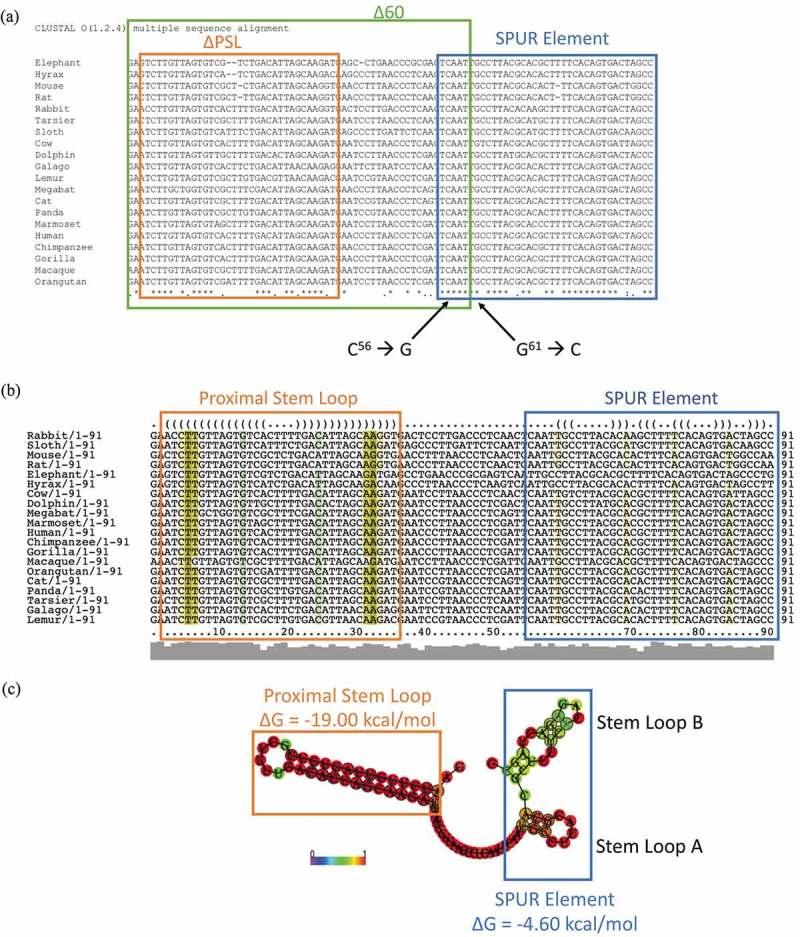
The proximal SelS 3′ UTR contains two conserved regions. a. The sequences of the first 91 nucleotides of SelS 3′ UTR from different mammals (Table S3) were analysed for nucleotide conservation by the ClustalOmega alignment program. Nucleotide positions with ‘*’ underneath have complete conservation across all species. Positions with ‘:’ have conservation between groups of strongly similar properties and positions marked with a ‘.’ have conservation between groups with weakly similar properties. The green box shows the first 60 nucleotides of the SelS 3′ UTR. The orange box shows the nucleotides that make up the PSL and the blue box encompasses the SPUR element. Nucleotides C56 and G61 were mutated to create the SPURdm. b. SelS 3′ UTR sequences were analysed for predicted structure using the RNAlifold program. The colour code indicates the number of base pair types found at each position: ochre-2, green-3, turquoise-4, blue-5, violet-6. Less saturated colours indicate that a base pair cannot be formed in some of the sequences. The orange box denotes the PSL and the blue box shows the SPUR element. c. The predicted consensus secondary structure of the first 91 nucleotides of the SelS 3′ UTR. Nucleotides shown in black circles indicate compensatory mutations within the sequences. The probability of a base pair is indicated on a scale from 0 (blue) to red (1) as shown in the colour bar. The PSL in orange has a highly conserved structure while the SPUR element (blue box) is made up of two smaller stem-loops. Free energies (ΔG) are shown for the PSL and the SPUR element.
