Figure 4.
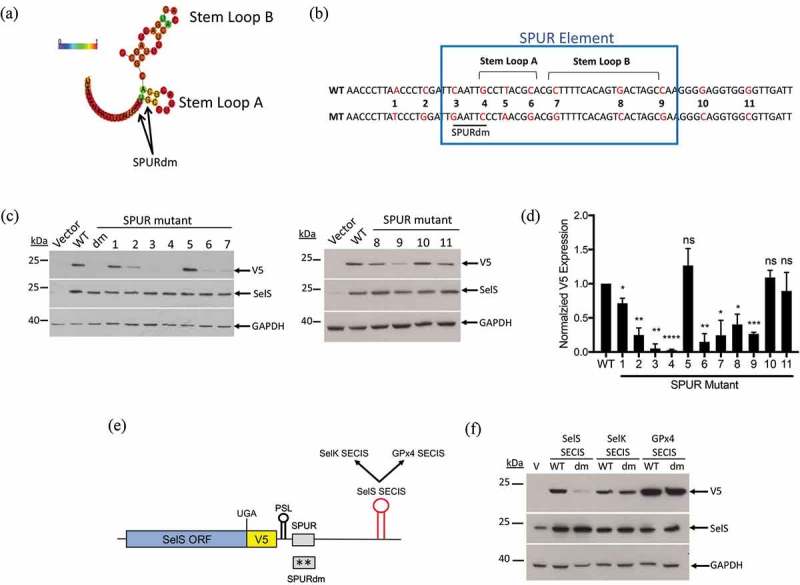
Requirements for SPUR element activity. a. Schematic showing the RNAlifold predicted structure of the human SPUR element. Nucleotide colours show probability of base pairing from 0 (blue) to 1 (red) as shown as described in the legend for Fig 3. The two nucleotides mutated in the SPURdm are indicated by arrows. b. Schematic of human SPUR element sequence. The blue box indicates the conserved nucleotides of the SPUR element. Stem Loops A and B are denoted by brackets. Red nucleotides in the sequence show the original nucleotide (WT) and what it was mutated to (MT). c. Representative Western blots showing the effect of single point mutations on V5 expression. Westerns were performed as described in Fig. 2b. d. Quantification of Western blots. V5 expression was quantified and normalized to SelS overexpression and GAPDH loading. Normalized V5 levels for each point mutant were compared to wild-type. Data represents results from two independent experiments analysed in duplicate (n = 4). *; p < 0.05, **; p < 0.01, ***; p < 0.001, ****; p < 0.0001, and ns – not significant). E. Schematic representation of the SelS-V5/UGA construct. The SelS SECIS element was replaced with the SECIS element from either SelK or GPx4. These SECIS elements were tested in both the wild-type SPUR and the SPURdm contexts. F. Representative Western blot from two independent experiments. WT = wild-type 3′ UTR, dm = SPURdm, and V = vector only. Western blots were performed as described in Fig 2(b).
