Figure 4: ReactomeFI Analysis of the Top Citrullinated Proteins in Heart Diseases.
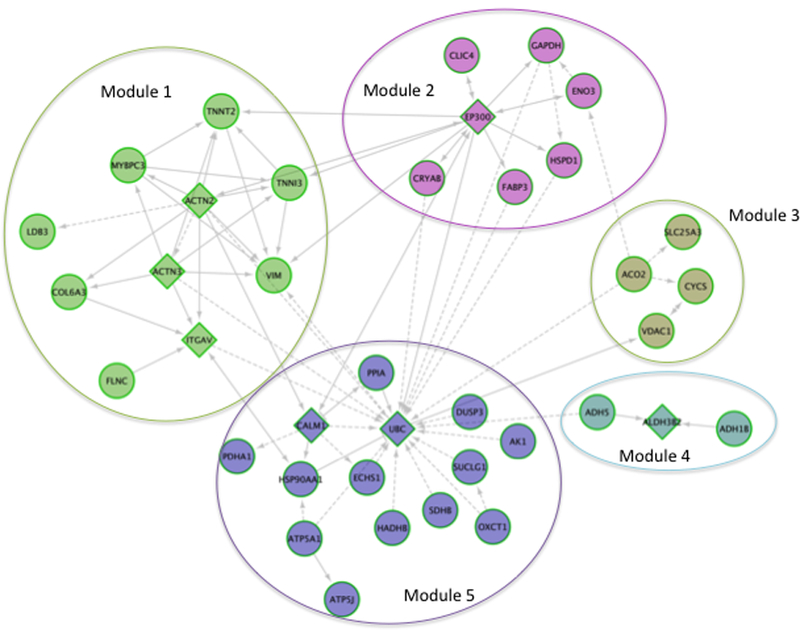
The top citrullinated proteins for [ref] were up-loaded and analyzed in cytoscape via reactomeFI. Circles nodes represent proteins that were differentially citrullinated, whereas triangle nodes represent proteins that were not reported as having differenitally citrullinated residues, but are linked to proteins, through protein-protein interactions (grey lines) that do have differentially regulated citrullinated residues. The top 3 pathways enriched per module were extracted. Module 1: Striated muscle contraction, hypertrophic cardiomyopathy, and dilated cardiomyopathy. Module 2: glycolysis/gluconeogenesis. Biosynthesis of amino acids, and validated targets of c-myc transcriptional activation. Module 3: Parkinson’s disease, The citric acid cycle and respiratory electron transport, and Huntington disease. Module 4: Tyrosine metabolism, Fatty acid degradation, and retinol metabolism. Module 5: The citric acid (TCA) cycle and respiratory electron transport, carbon metabolism, and metabolic pathway.
