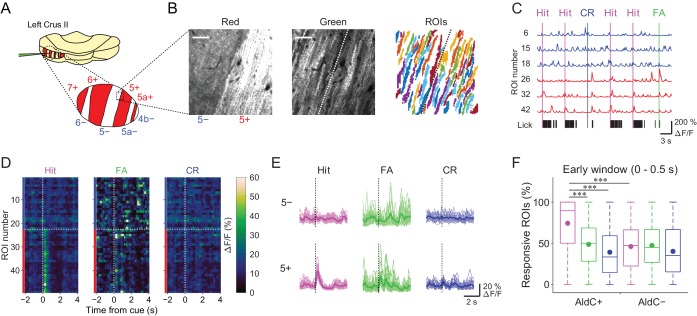
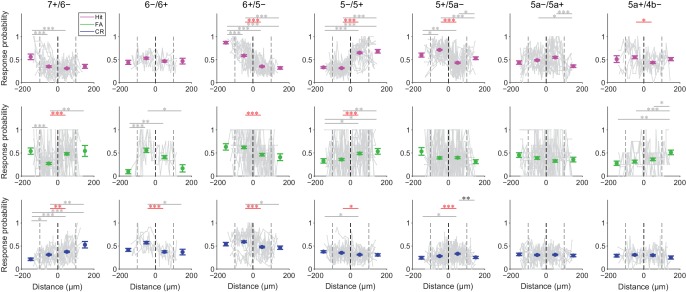
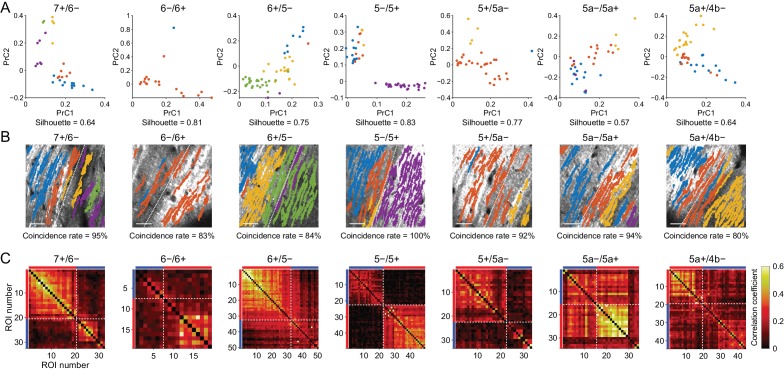
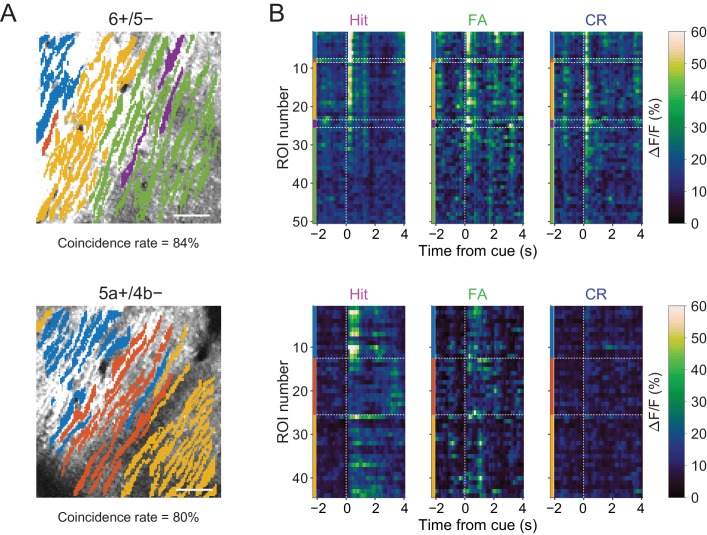
Figure 2. Differences in task-related CF signals between AldC+ and AldC− compartments.
(A) Schematic diagram of virus injection into the left cerebellar folium Crus II of Aldoc-tdTomato mice. Inset, magnified view of the Crus II. Red stripes represent AldC+ compartments. (B) Left, tdTomato fluorescence image at a 5−/5+ boundary. Middle, mean image of a GCaMP6f video at the same field of view. Right, extracted ROIs representing PC dendrites that are pseudocolored, in the same field of view. Scale bars, 40 μm. (C) Representative calcium traces from dendrites in panel (B) and their corresponding lick activities (black bars). Colored vertical lines represent the onsets of sensory cues for each trial type. (D) Trial-averaged calcium traces of single ROIs from the data in panel (C). (E) Single ROI trial-averaged traces for 5− and 5+ compartments from the data in panel (C). Thick lines represent ROI-averaged traces. (F) Percentage of responsive ROIs in a compartment per session for each trial type pooled across all AldC+ or AldC− compartments (n = 17 mice, 79 imaging sessions). Colored circles represent means. Colors represent trial types (hit, magenta; FA, green; CR, blue). Two-way ANOVA on ranks with repeated measures followed by post-hoc Tukey’s test: ***p<0.001.