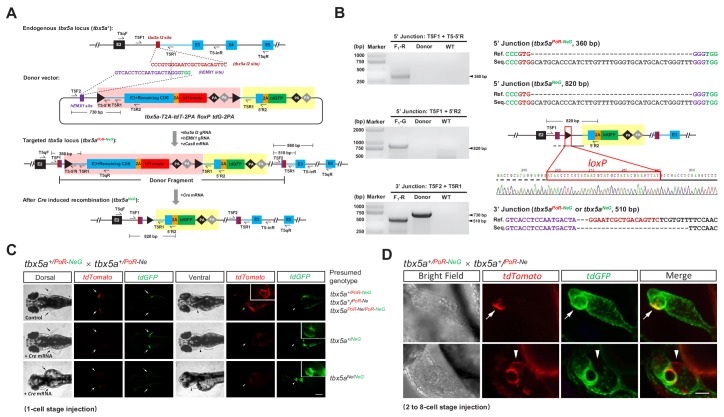
Figure 3. Generation of geno-tagging alleles by improving the dual-cassette donor strategy at the zebrafish tbx5a locus.
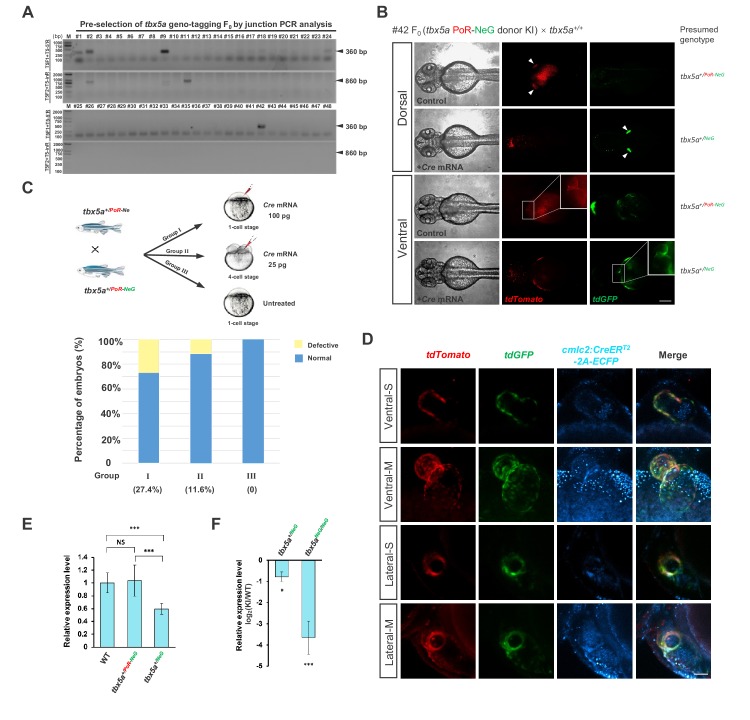
(A) Schematic diagram of the KI strategy for geno-tagging based on the tbx5a-T2A-tdT-2PA floxP tdG-2PA donor (or abbreviated as tbx5a PoR-NeG donor). Primers T5qF and T5qR are used for qRT-PCR in Figure 3—figure supplement 1E and F. (B) Results of junction PCR and direct sequencing to detect the tbx5a geno-tagging donor knockin and Cre-induced recombination events in 48 hpf embryos obtained from the cross in Figure 3—figure supplement 1B, that is the F1 embryos from the #42 positive F0 outcrossed with a wild-type zebrafish before or after Cre mRNA injection. Note that the sequences of both the T5F2 and T5R1 primers are also present in the donor vector, flanking the upstream loxP site (as shown in panel A); therefore, a 730 bp product could be amplified in the lane with the donor as the template. F1-R: an F1 embryo showing a red fluorescent signal (before Cre mRNA injection). F1-G: an F1 embryo showing a green fluorescent signal (after Cre mRNA injection). Donor: tbx5a PoR-NeG geno-tagging donor plasmid. WT: pooled genomic DNA of five wild-type embryos. (C) Phenotype analysis of the 72 hpf embryos from tbx5a+/PoR-NeG heterozygotes (derived from F0 #42 in Supplementary file 5) crossed with a tbx5a+/PoR-Ne heterozygote (derived from F0 #12 in Supplementary file 3) after the injection of Cre mRNA at the 1 cell stage. The upper panel represents an uninjected control embryo showing only a red fluorescent signal, whose genotype should be either tbx5a+/PoR-Ne, tbx5a+/PoR-NeG or tbx5a PoR-Ne/PoR-NeG. The middle panel represents a Cre mRNA-injected embryo showing normal development, whose genotype is expected to be tbx5a+/NeG. The lower panel represents a Cre mRNA-injected embryo showing a typical tbx5a mutant phenotype, including heart region defects and a lack of pectoral fins, whose genotype is expected to be tbx5aNe/NeG. Arrows indicate the pectoral fins, and arrowheads indicate the heart region. The boxed insets show a higher magnification of the corresponding heart region, for better comparison of heart morphology. Scale bar, 200 μm. (D) Z-stack confocal images of two representative 48 hpf embryos after the injection of Cre mRNA at the 2- to 8 cell stage from the same cross as in C. The white arrows indicate the colocalization of the tdGFP and tdTomato signals, and the white arrowheads indicate the mutually exclusive expression of the tdGFP and tdTomato signals. Scale bar, 50 μm.