Figure 3.
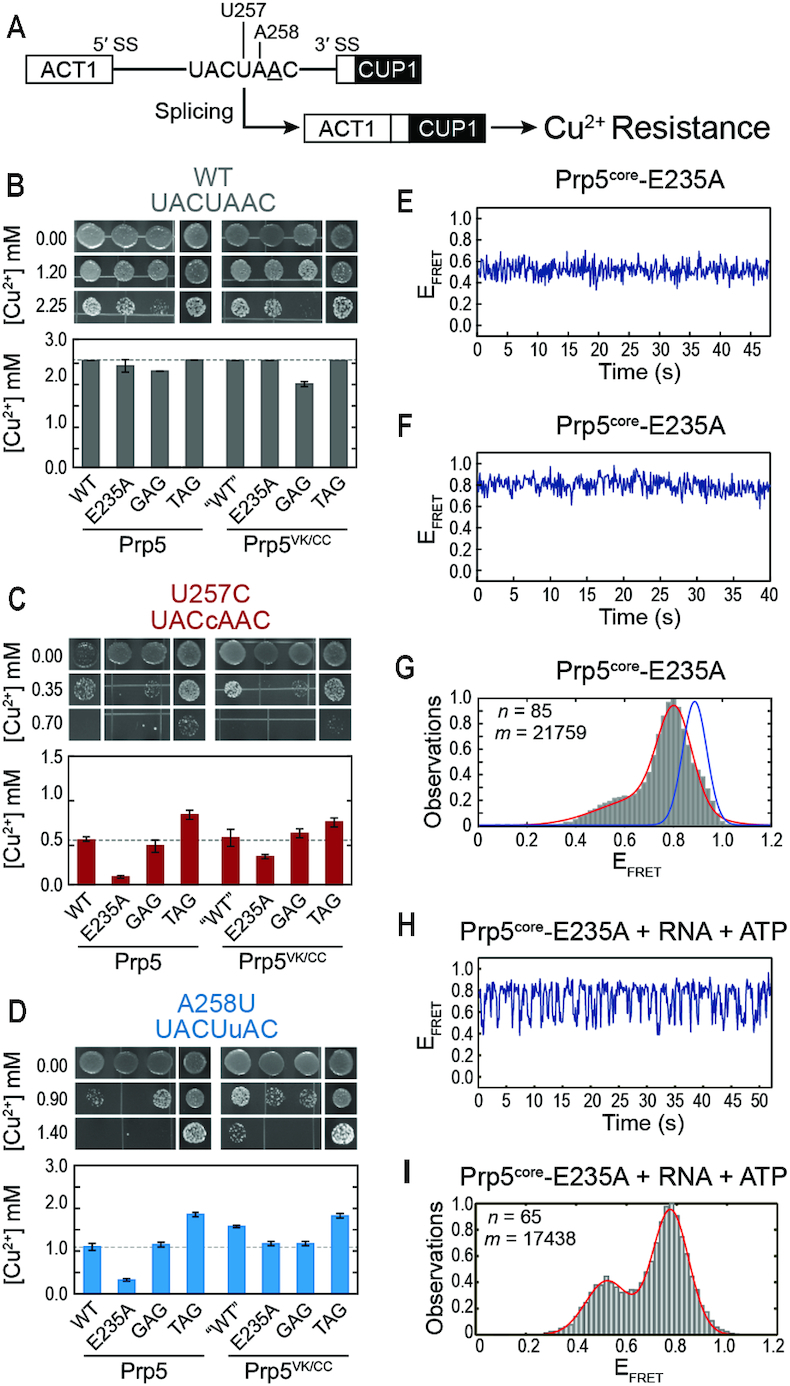
The Prp5-E235A substitution increases splicing fidelity in yeast and conformational dynamics of Prp5core. (A) Schematic of the ACT1-CUP1 reporter pre-mRNA and assay. The BP-A is underlined, and positions of BS mutations are noted. (B–D) Results from Cu2+ growth assays of strains containing the indicated Prp5 mutations and either the consensus, U257C, or A258U ACT1-CUP1 reporter plasmids. Results are the average of quadruplicate experiments ± the standard deviation. (E, F) Representative EFRET trajectories for two molecules of Prp5core-E235A (G) Histogram of EFRET values obtained for Prp5core-E235A (grey boxes) and a fit of the data (red line). Blue line is from Figure 2C. (H) Representative calculated EFRET trajectory for a single molecule of Prp5core-E235A in the presence of poly(A) RNA and ATP. (I) Histogram of EFRET values obtained in the presence of poly(A) RNA and ATP as in panel (G). n and m represent the number of individual molecules and EFRET data points, respectively, used to construct the histogram. Data fit results are listed in Supplementary Table S4.
