Figure 7.
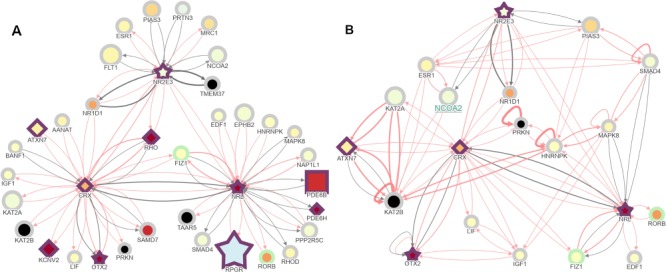
A control case interaction visualized on RPGeNet showing the connections between NRL, NR2E3 and CRX retinal transcription factors. (A) RPGeNet was queried to display the subnetwork among NRL, NR2E3 for the nodes at distance one at level one; then nodes connected to CRX were added with the ‘Node Addition’ button activated on click. CRX, NRL and NR2E3 are three well-known transcription factors that co-regulate retinal-specific genes, among them RHO. Interestingly, several other genes that cause retinal dystrophies also appear in the network as target genes or other transcriptional regulators (border shown in purple). Nodes color-fill defined by the ‘RET-ALL’ gene-expression data. (B) Further trimming of the subnetwork obtained by omitting the nodes that are not chromatin remodelers or transcription factors provide an overview of relevant co-regulators of retinal genes. After deleting the corresponding nodes, further edges—out of the shortest paths that link the genes left—were shown by clicking on the ‘Connect Genes’ button at the control panel. This visualization allows pinpointing and exploring alternate pathways that connect the initial seeds; for instance, CRX and NRL were already connected, but longer paths are now evident like CRX⇌PRKN⇌HNRNPK⇌NRL, when PRKN and HNRNPK are linked. Another example can be the pathway found between NRL and NR2E3 on the path NRL⇌SMAD4⇌PIAS3⇌NR2E3, when SMAD4 and PIAS3 are linked. Both panels from this figure can be reproduced on the Network Explorer if users upload the Supplementary Files 2 and 3, respectively.
