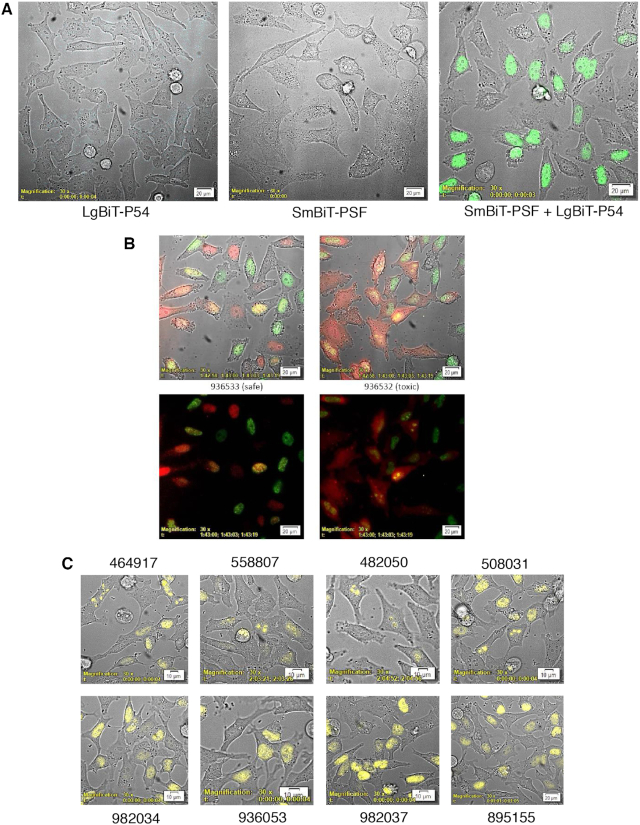
Figure 2.
P54nrb-PSF NanoBiT assay. (A) Evaluation of LgBiT-P54nrb expression in CRISPR cell line. HeLa CRISPR LgBiT-P54nrb or HeLa control cells seeded on glass-bottom culture dishes then transfected with a plasmid expressing a SmBiT-PSF fusion protein (pSmB-PSF). The following day NanoGlo substrate was added to the cells and luminescence visualized using an Olympus LV200 Inverted microscope. Left panel, HeLa LgBiT–P54 fusion; center, control HeLa cells transfected with pSmB-PSF; right, HeLa LgB-P54 cells transfected with pSmB-PSF. (B) Safe/Toxic PS-ASO association with P54nrb-PSF heterodimer in live cells. The LgBiT-P54nrb HeLa cell line was transfected with pSmBiT-PSF. The following day cells were treated with Alexa595-linked safe ASO 936533 (left panels) or toxic ASO 936532(right panels) at 100 nM. Cells were then visualized using a bioluminescent imaging microscope at 3-min intervals for a total of 2.5 h collecting brightfield, ASO fluorescence (red) and NLuc bioluminescence (green). Lower panels; brightfield removed. This figure is representative of numerous fields and was repeated 3–4 times. For videos see Supplementary Figure S5B/C. (C) Substitution with 2′-methoxy nucleotide at position 2 of the DNA ‘gap’ portion of toxic PS-ASOs prevents nucleolar accumulation of P54nrb/PSF heterodimer. The LgBiT-P54nrb HeLa cell line was transfected with pSmBiT-PSF. The following day cells were treated at 200 nM for 2.5 h with toxic PS-ASOs (upper panels) or ASOs of the same sequence substituted at gap position 2 with 2′-methoxy (lower panels). Cells were then visualized using a bioluminescent imaging microscope collecting brightfield and NLuc bioluminescence (yellow).