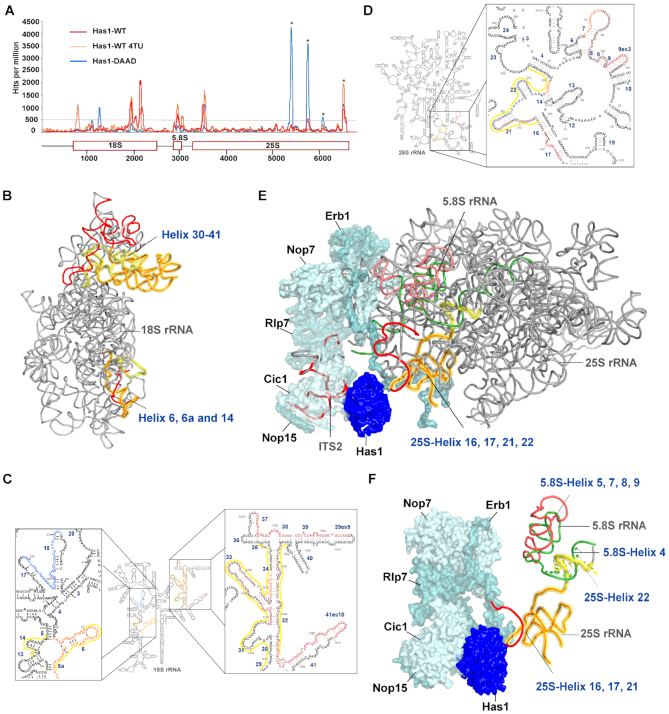
Figure 3.
Has1 crosslinks to multiple regions of pre-rRNAs. (A) Plot showing the distribution of reads coverage across pre-rRNA. Average number of hits from two independent experiments was plotted. Reads per 1 million total reads are shown. Asterisks indicate common contaminating peaks. (B) Binding sites of Has1-WT highlighted in 3D structure of mature 40S (PDB:4V88). (C) and (D) Crosslink sites of both Has1-WT and DAAD mutant highlighted in the secondary structure of 18S or 5.8S and 25S rRNAs obtained from RiboVision suite (47): red - Has1 wild type, orange - Has1 wild type 4TU, blue - Has1 DAAD. The overlapping regions of Has1 wild type from standard and 4TU CRAC are shown in dark red. The overlapping regions of Has1 wild type and Has1-DAAD are in dark blue. Has1 binding sites obtained from Brüning et al., (21) are highlighted by a yellow line next to the sequence. (E) Crosslink sites of Has1-WT mapped on the 3D structure of pre-60S ribosome (PDB:6EM5). (F) Detail of the 5.8S/25S crosslink in the 3D structure of pre-60S ribosome (PDB:6EM1). In the 3D structures, Has1 wild type crosslinked regions are shown in red. The crosslinking regions identified by Brüning et al. are highlighted in yellow, with the overlap in orange. The 5.8S rRNA is shown in green and the crosslinked region is in salmon.