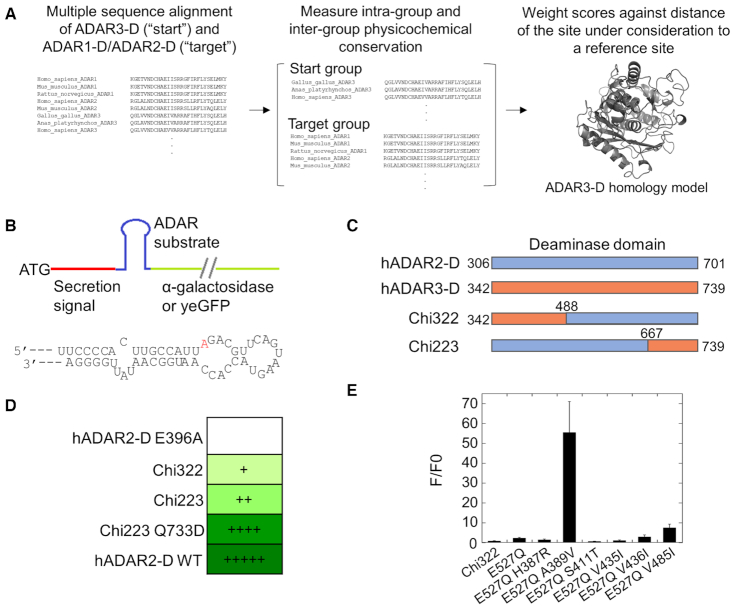
Figure 1.
Janus prediction and hADAR2-D/hADAR3-D chimeras for testing the importance of mutations predicted by Janus. (A) Workflow of Janus program for predicting mutations important for ADAR3 activation. (B) Top: Scheme of the reporters for testing A-to-I editing activity (26,27). The colorimetric reporter and the fluorescent reporter use α-galactosidase and yeGFP (yeast enhanced green fluorescent protein) as the reporter protein, respectively. The secretion signal sequence exists only in the colorimetric reporter but not the fluorescent reporter. Bottom: sequence of the ADAR substrate used in the reporters. The editing site is colored in red. (C) Scheme of composition of hADAR2-D/hADAR3-D chimeras. The numberings in the chimeras are based on hADAR3 sequence. (D) A-to-I editing activity of the chimeras evaluated with the colorimetric reporter assay. ‘+’ indicates that yeast colonies started turning green after >30 days of incubation; ‘++’ indicates 9 days; ‘++++’ indicates 4 days; ‘+++++’ indicates 2 days. (E) The effect of mutations predicted by Janus on Chi322 activity using the fluorescent reporter assay. Mutations were introduced into Chi322. F/F0 is the ratio of fluorescence of cells expressing each mutant construct divided by fluorescence of cells expressing hADAR2-D inactive mutant E396A. Error bar indicates SD, n ≥ 3.