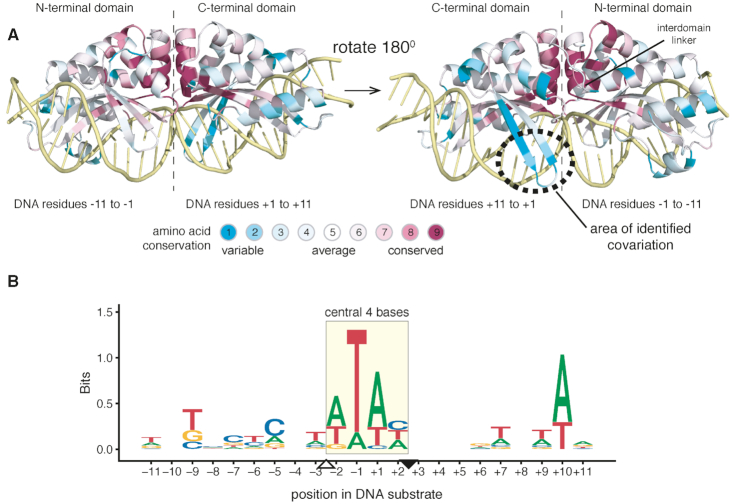
Figure 1.
Overview of LAGLIDADG structure and evolutionary conservation. (A) Evolutionary conservation of LAGLIDADG homing endonucleases overlaid on the structure of the monomeric I-OnuI (modified from PDB ID: 3QQY). The N- and C-terminal domains of I-OnuI are indicated, as are the (−) and (+) halves of the DNA substrate. The color scheme is based on a conservation score generated by the ConSurf program, with variable residue positions given a score of 1 and highly conserved residue positions a score of 9 (55). (B) Logos alignment of aligned DNA target sites used in the MI analysis. The substrates are labeled according to standard nomenclature with the central 4 bases indicated by a rectangle and the top- and bottom-nicking sites indicated by downward and upward facing rectangles, respectively.