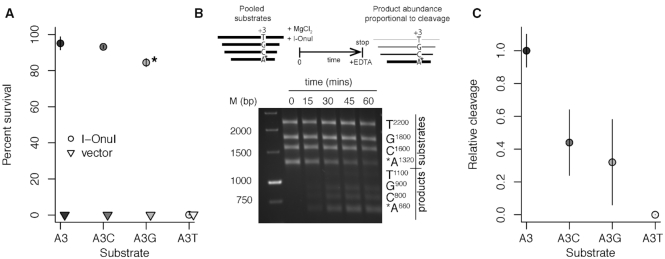
Figure 3.
Nucleotide substitutions at position +3 impact I-OnuI in vivo and in vitro activity. (A) I-OnuI in vivo survival on +3 substrates tested in an Escherichia coli two-plasmid selection system. Circles represent mean survival values from at least three independent experiments with I-OnuI, and inverted triangles mean survival values from from at least three independent experiments with the vector. The lines represent the range of error. Symbols are shaded by substrate identity. The asterisk (*) indicates a slow-growth colony phenotype associated with the A+3G substrate. (B) Top, schematic of in vitro barcoding assay where substrates of different lengths contain nucleotide substitutions in the +3 position. Incubation over time generates products of different lengths. Bottom, representative cleavage assay analyzed by gel electrophoresis in 0.8% agarose with lengths of +3 substrates and products indicated. (C) Relative cleavage of the +3 substrates derived from quantitative analysis of product and substrate ratios from panel (B). Circles indicate the mean relative cleavage from at least 3 independent experiments and the lines represent the range of error.