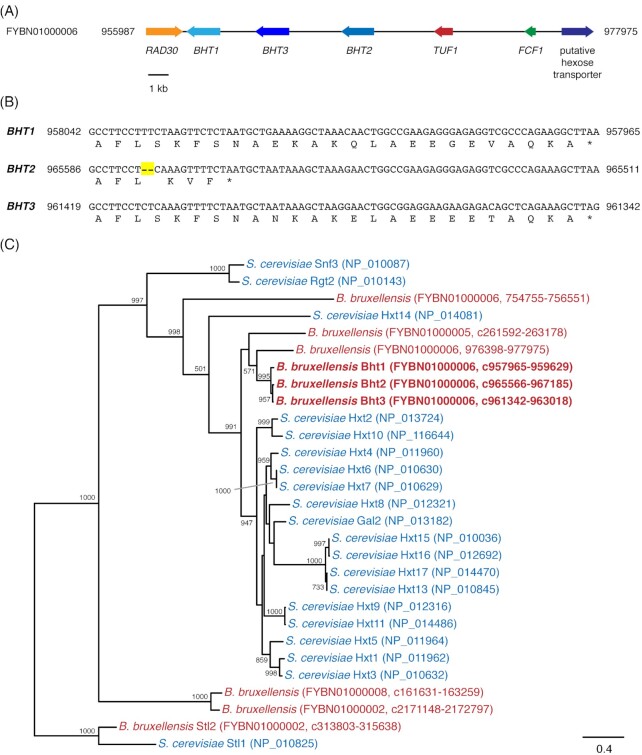
Figure 1.
Identification and characterization of hypothetical genes encoding putative glucose transporters in the genome of B. bruxellensis. (A), Genomic context of the BHT1-BHT3 gene cluster in the genome of B. bruxellensis UMY321. Gene names of adjacent genes were assigned based on sequence homology to previously described genes in S. cerevisiae. (B), Alignment of the 3’ termini of the B. bruxellensis hypothetical genes BHT1, BHT2 and BHT3. The proposed 2-bp deletion in the BHT2 sequence is indicated in yellow. Genomic coordinates of the source contig (GenBank accession FYBN01000006) are indicated. (C), Maximum likelihood tree of predicted glucose transporters in S. cerevisiae strain S288C (blue) and B. bruxellensis strain UMY321 (red) using 423 aligned amino acid positions. Node stability is indicated by the bootstrap value of 1000 replicate analyses. Bootstrap values below 500 are not shown. The S. cerevisiae glycerol transporter Stl1p and its B. bruxellensis ortholog (Zemancíková et al. 2018) were used as outgroups. GenBank protein accession numbers for S. cerevisiae transporters are displayed. For B. bruxellensis hypothetical genes, the genomic contig GenBank accession number and genomic coordinates used for conceptual translation are indicated (‘c’ signifies that the hypothetical gene is encoded on the reverse strand). The three B. bruxellensis putative glucose transporters BHT1-BHT3 are highlighted in bold font.