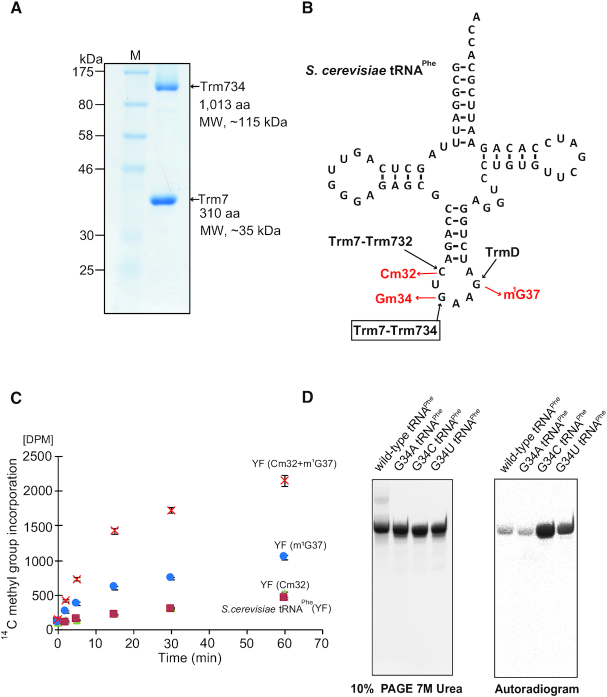
Figure 1.
Characterization of the recombinant Trm7–Trm734 (A) SDS-PAGE (12.5%) of purified recombinant Trm7–Trm734. The gel was stained with Coomassie Brilliant Blue. (B) Cloverleaf structure of Saccharomyces cerevisiae tRNAPhe transcript. Three SAM-dependent tRNA methyltransferases, Trm7–Trm732, Trm7–Trm734 and TrmD, catalyze formation of Cm32, Gm34 and m1G37, respectively. (C) Time-dependent methyl transfer activity of Trm7–Trm734 using S. cerevisiae tRNAPhe transcript (YF, green triangles), YF with Cm32 (red squares), YF with m1G37 (cyan circles) and YF with both Cm32 and m1G37 (orange cross marks). Error-bars indicate the standard deviation calculated from the results of three independent experiments. (D) Methyl-group acceptance activities of mutant tRNAPhe transcripts were measured. G34 in the wild-type transcript was replaced by A, U or C. Left, the transcripts were analyzed by 10% PAGE (7 M urea). The gel was stained with methylene blue. Right, autoradiogram of the same gel.