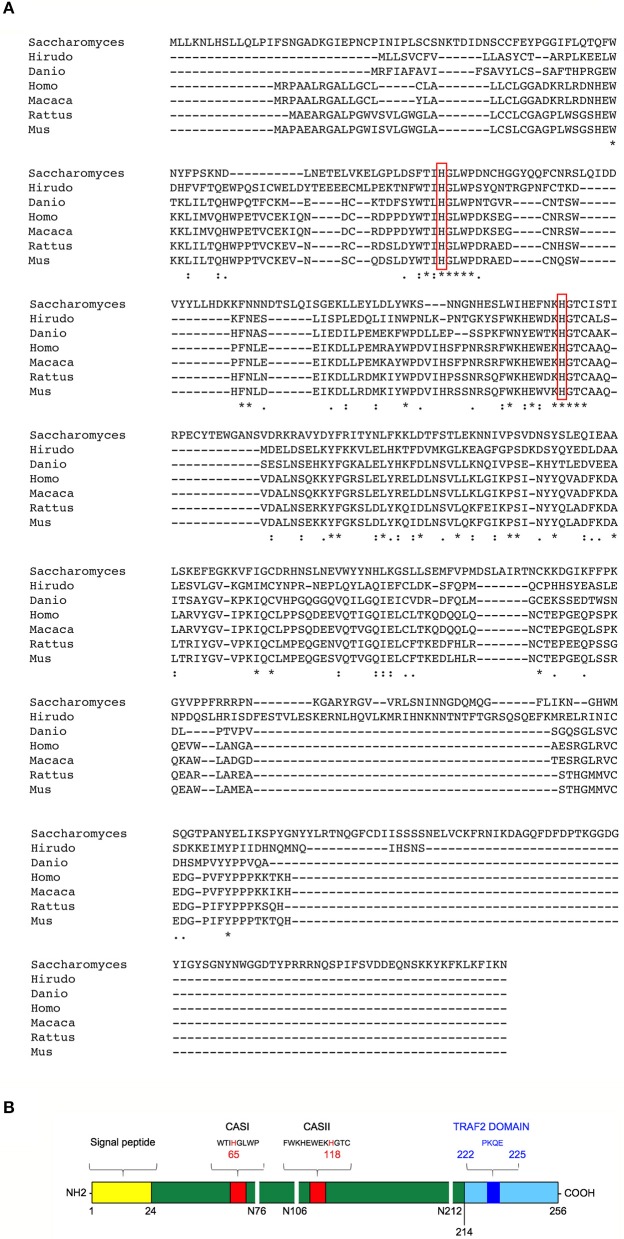
Figure 1.
Structural features and evolutionary conservation of T2 RNase proteins. (A) A Clustal Omega alignment of several members of the T2 RNase family, showing the wide evolutionary conservation of this enzymes. RNA cleavage by T2 RNases is mediated by histidine residues (red boxes) embedded into two highly conserved motifs dubbed CAS I and CAS II. The species included in the alignment are Saccharomices cerevisiae, Hirudo verbana, Danio rerio, Homo sapiens, Macaca mulatta, Rattus norvegicus, and Mus musculus. (B) Structure of human RNASET2. The RNASET2 primary sequence includes 256 aminoacid residues, with a predicted molecular weight of about 30 kDa. The core enzyme is colored in dark green in the figure. Among the protein's structural features, a 24-residues long signal peptide for secretion at the N-terminal (yellow bar) and the two canonical CAS sites (CAS I/II, red bars) responsible for the enzyme's catalytic activity (bearing a highly conserved key histidine residue at position 65 and 118, respectively) are shown in the figure. Three N-glycosylation sites (white horizontal lines), which increase the molecular weight of the native protein of about 6 kDa, are also shown (34). Finally, a putative TRAF-2 binding site (dark blue bar) was predicted in the C-terminal part of RNASET2 (light blue bar, starting form residue 214), which is less evolutionary conserved throughout evolution and has been suggested to be comprise a highly disordered loop (35). Within the cell, the RNASET2 protein is present in three forms of different sizes, namely 36, 31, and 27 kDa (34). The 36 kDa isoform represents the full-length and secreted form, which is easily detected in cell culture supernatants from RNASET2-expressing cells, whereas the other two isoforms represent intracellular protein isoforms originating from proteolytic cleavage of the full-length protein (B).