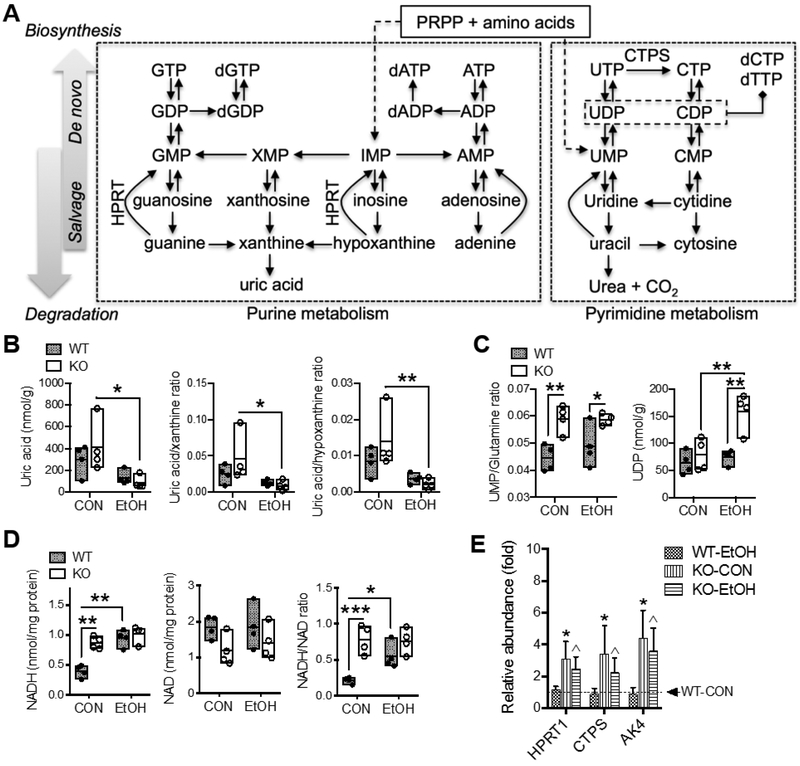
Fig. 6. Alterations in metabolites and gene expression involved in nucleic acids metabolism and nucleotide cofactors.
(A) Scheme of selected biochemical reactions and metabolites involved in purine and pyrimidine metabolism. Solid or dashed lines with arrow indicate single or multiple biochemical reactions, respectively. Hepatic concentrations of metabolites of (B) purine metabolism and (C) pyrimidine metabolism; results are expressed in mole per gram (g) liver weight. (D) Hepatic concentrations of NAD and NADH; results are expressed in mole per milligram (mg) total proteins. All data are presented as floating bars (min to max, line at mean; N = 4/group) showing individual data points from wild-type (WT, closed circles) and Gclm-KO (KO, open circles) mice. *P < 0.05, **P < 0.01, ***P < 0.001 by two-way ANOVA with post-hoc Holm-Sidak's multiple comparisons test. (E) Relative mRNA abundance measured by Q-PCR. Results are expressed as mean ± S.D. (N = 4/group). *Compared to WT-CON, ^compared to WT-EtOH, or #compared to KO-CON: P < 0.05 by two-way ANOVA with post-hoc Holm-Sidak's multiple comparisons test. WT-CON, wild-type mice fed control diet; KO-CON, Glcm KO mice fed control diet; WT-EtOH, wild-type mice fed ethanol-containing diet; KO-EtOH, Glcm KO mice fed ethanol-containing diet; AK4, Adenylate kinase 4; CTPS, CTP synthase; HPRT, hypoxanthine phosphoribosyltransferase. PRPP, 5-phosphoribosyl-1-pyrophosphate