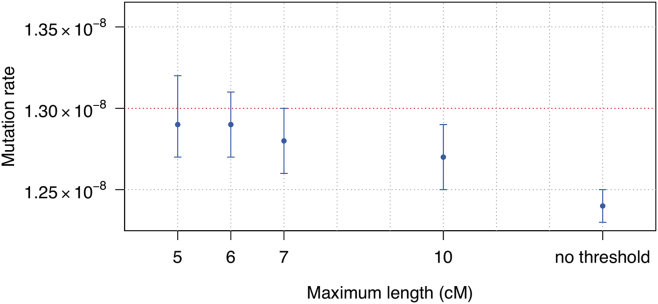
Figure 4.
Estimated Mutation Rates under False Negative Genotype Errors
Data were simulated under the “homogeneous” model, and errors were added using the “false-negative” genotype error scheme (described in Material and Methods). The simulated mutation rate is 1.30 × 10−8 per base pair per meiosis and is indicated with the red dotted line. We set different thresholds on the maximum length of IBD segments (x axis) to control the impact of false negative errors. The maximum allele frequency threshold for the variants used for the mutation rate estimation is 3.75%. Point estimates (dots) and 95% confidence intervals (bars) are shown.