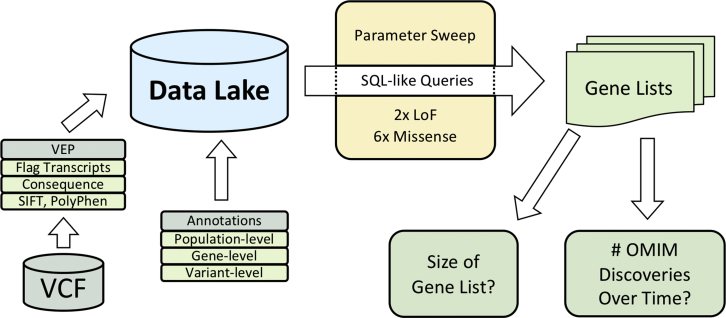
Figure 1.
HARLEE Workflow
ES VCF files are first annotated with Variant Effect Predictor (VEP), where one transcript is flagged per variant per gene. Consequence, SIFT, PolyPhen, variant allele frequency from multiple sources, domain information, and other annotations are additionally ascertained by VEP. VEP output is loaded into a Hadoop architecture data lake. Finally, population-, variant- and gene-level annotations from a variety of sources are loaded, allowing for modular, on-demand annotation. After samples and annotations are separately loaded into HARLEE, a series of SQL-like queries generate distinct gene lists. Bioinformatic filtering parameters based on loaded annotations are tuned to optimize discovery density, which takes into account the volume of genes reported to OMIM as disease-associated over time normalized against the number of remaining genes without OMIM disease annotations.