Abstract
The remarkable pro‐apoptotic properties of tumour necrosis factor (TNF)‐related apoptosis‐inducing ligand (TRAIL) have led to considerable interest in this protein as a potential anticancer therapeutic. However, TRAIL is largely ineffective in inducing apoptosis in certain cancer cells, and the mechanisms underlying this selectivity are unknown. In colon adenocarcinomas, posttranslational modifications including O‐ and N‐ glycosylation of death receptors were found to correlate with TRAIL‐induced apoptosis. Additionally, mRNA levels of fucosyltransferase 3 (FUT3) and 6 (FUT6) were found to be high in the TRAIL‐sensitive colon adenocarcinoma cell line COLO 205. In this study, we use agonistic receptor‐specific TRAIL variants to dissect the contribution of FUT3 and FUT6‐mediated fucosylation to TRAIL‐induced apoptosis via its two death receptors, DR4 and DR5. Triggering of apoptosis by TRAIL revealed that the low FUT3/6‐expressing cells DLD‐1 and HCT 116 are insensitive to DR5 but not to DR4‐mediated apoptosis. By contrast, efficient apoptosis is mediated via both receptors in high FUT3/6‐expressing COLO 205 cells. The reconstitution of FUT3/6 expression in DR5‐resistant cells completely restored TRAIL sensitivity via this receptor, while only marginally enhancing apoptosis via DR4 at lower TRAIL concentrations. Interestingly, we observed that induction of the salvage pathway by external administration of l‐fucose restores DR5‐mediated apoptosis in both DLD‐1 and HCT 116 cells. Finally, we show that fucosylation influences the ligand‐independent receptor association that leads to increased death inducing signalling complex (DISC) formation and caspase‐8 activation. Taken together, these results provide evidence for the differential impact of fucosylation on signalling via DR4 or DR5. These findings provide novel opportunities to enhance TRAIL sensitivity in colon adenocarcinoma cells that are highly resistant to DR5‐mediated apoptosis.
Keywords: death receptor 5, fucosylation, glycosylation, receptor clustering, TNF‐related apoptosis‐inducing ligand
Because of its ability to induce apoptosis in tumour cells, TRAIL [Tumour necrosis factor (TNF)‐related apoptosis‐inducing ligand] has sparked a lot of interest in oncology research. Posttranslational modifications and upregulation of fucosylation were previously found to correlate with TRAIL‐induced apoptosis. This study shows that fucosylation triggers the clustering of death receptors DR4 and DR5, leading to increased cell death. Moreover, apoptosis mediated by DR5 can be stimulated by simple external administration of l‐fucose.
Abbreviations
- 2FF
2F‐peracetyl‐fucose
- bGalNAc
benzyl 2‐acetamido‐2‐deoxy‐α‐d‐galactopyranoside
- DISC
death inducing signalling complex
- DR4
death receptor 4
- DR5
death receptor 5
- FUT3
fucosyltransferase 3
- FUT6
fucosyltransferase 6
- l‐fucose
l‐(‐)‐Fucose
- TRAIL
TNF‐related apoptosis‐inducing ligand
Introduction
Colorectal cancer is the third most commonly diagnosed cancer in males and the second in females as estimated by the International Agency for Research on Cancer in 2018 1. Although early detection methods, increased awareness and improved treatment modalities have led to a reduction in both incidence and cancer death rates in several western countries, colorectal neoplasia are still one of the most deadly cancers, causing around 880 000 deaths worldwide every year 1. The 5‐year survival rate of colorectal cancer patients for localized‐stage is 90%, but decreases to 71% and 14% for regional and distant stage, respectively 2, although some relatively new strategies are developed to predict surgical resection more accurately such as ‘watch and wait’ strategy and Narrow‐band Imaging International Colorectal Endoscopic Classification (NICE) 3, 4. Thus, novel therapies are required to improve the prognosis of colorectal cancer patients.
The TNF superfamily member TRAIL has gathered considerable attention as a potential cancer therapeutic, as it is able to induce apoptosis selectively in tumour cells while leaving normal cells unharmed 5, 6, 7. TRAIL is capable of signalling via two apoptosis‐inducing transmembrane death receptors (DRs), named DR4 (TRAIL‐R1) and DR5 (TRAIL‐R2). TRAIL also binds to three decoy receptors: DcR1 (TRAIL‐R3), DcR2 (TRAIL‐R4) and the soluble decoy receptor osteoprotegerin (OPG) 8, 9. Decoy receptors are unable to transduce death‐inducing signals as they lack a functional intracellular death domain (DD) 9, 10, 11, 12, and they can diminish apoptosis activation by competing with TRAIL‐DR interactions 10, 13 or by forming nonsignalling heterotrimeric complexes 14.
Binding of trimeric recombinant human TRAIL (rhTRAIL) to death receptors triggers the intracellular formation of the so‐called death inducing signalling complex (DISC), consisting of Fas‐associated death domain (FADD), which further recruits and activates pro‐caspase‐8 and/or pro‐caspase‐10 15, 16, 17, 18. The activation of initiator caspases leads to direct cleavage and activation of executioner caspases‐3 and ‐7 and subsequent apoptosis induction through the extrinsic apoptotic pathway 19, 20, 21, 22. Activated caspases also lead to the cleavage of Bid (tBid), resulting in the release of mitochondrial factors, cleavage of caspase‐9 and activation of effector caspases leading to apoptosis via the intrinsic pathway 23. Despite its reported tumour selective properties, several studies showed that approximately 50% of the colorectal cancer cell lines are resistant to rhTRAIL 24, 25. Untagged soluble human TRAIL Dulanermin developed for clinical use was shown to have a half‐life of only 23–31 min in nonhuman primates in preclinical tests, which was attributed to its low mass 26. In order to improve half‐life some TRAIL‐R1/R2 agonist antibodies have been developed, which have a higher mass and therefore a prolonged half‐life. However, Mapatumumab (HGS‐ETR1), TRAIL‐R1 agonistic antibody, in Phase II clinical trial did not show satisfactory activity with refractory colorectal cancer patients 27. This was shown due to the resistance of colorectal cancer cell lines to TRAIL 24, 25, 28. The current focus is to use the combination strategies to overcome TRAIL resistance. One Phase Ib open‐labelled clinical study (NCT00671372) reported that the combination of Dulanermin and FOLFIRI greatly prolonged the patient's survival 29. The TRAIL‐R2 agonistic antibody, Conatumumab (AMG655) has been shown to be well‐tolerated (NCT00819169) in various cancers including advanced colorectal cancer 30. This clinical study is now continued to Phase II in combination with chemotherapeutic agents (NCT01327612).
Fucosylation is an important type of posttranslational modification in colon cancer 31, in which fucose residues are terminally attached to N‐ or O‐linked glycans or glycolipids 32, 33. This modification on the cell‐surface is essential in numerous biological processes, such as ontogeny, cellular differentiation and signalling events 32, 34. All fucosyltransferases require the donor substrate GDP‐fucose, which can be synthesized from GDP‐mannose by the dominant de novo pathway or from free l‐fucose by the salvage pathway 31, 33, 34. In the past years, several reports have described the importance of fucosylation in TRAIL‐induced apoptosis in colon cancer. Wagner et al. 24 firstly described the positive correlation between TRAIL sensitivity and mRNA levels of fucosyltransferase enzymes FUT3 and FUT6 in a panel of colon adenocarcinoma cells. Furthermore, mutation of the GDP‐mannose‐4,6‐dehydratase (GMDS) gene leads to the inactivation of the de novo GDP‐fucose pathway and decreased TRAIL sensitivity, resulting in accelerated tumour growth in vivo, due to a lack of NK cell‐mediated tumour surveillance 23. GMDS also plays an important role in the formation of the FADD‐dependent complex II, which comprises FADD, caspase‐8 and c‐FLIP. GMDS deficiency inhibited both DR4‐ and DR5‐mediated apoptosis by inhibiting the formation of the complex II, while it did not affect formation of the DISC or recruitment to and activation of caspase‐8 35. The same group showed that fucosylation could be regulated through DNA methylation. Treatment with the novel methyltransferase inhibitor Zebularine was found to increase fucosylation levels, leading to enhanced TRAIL‐induced apoptosis without increasing TRAIL receptor and/or caspase‐8 levels 36. However, it is still unclear whether the fucosylation of DR4 and DR5 equally contributes to TRAIL‐mediated apoptosis. Recently receptor specific agonists developed by us in the past have been used to unravel the respective contribution of DR4 and DR5 N‐glycosylation on TRAIL signalling 37, 38.
Here we investigated the more precise role of fucosylation on DR4‐ and DR5‐mediated apoptosis in colon adenocarcinomas, using TRAIL receptor‐specific apoptosis‐inducing variants that bind selectively and with high affinity to either DR4 or DR5 38, 39, 40, 41, 42. We show that fucosylation of DR4 and DR5, either via the salvage or via the de novo synthesis pathway, enhances TRAIL signalling in colon adenocarcinoma cells. We were able to increase DR5‐mediated apoptosis in DR5 resistant colon cancer cell lines by improving the fucosylation status of the death receptor.
Results
Variation in sensitivity to DR4‐ and DR5‐mediated apoptosis among different colon adenocarcinomas
To identify the sensitivity of colon adenocarcinoma cells to TRAIL via either DR4 or DR5, we investigated three cell lines: COLO 205, DLD‐1 and HCT 116. By detecting the Annexin V levels induced by WT TRAIL, the DR4‐specific TRAIL variant 4C7 and the DR5‐specific TRAIL variant DHER, we found that COLO 205 was highly sensitive to TRAIL‐mediated cell death via both death receptors (Fig. 1A). Cell death induction in DLD‐1 and HCT 116 cells is primarily mediated by DR4 and not by DR5 as evidenced by the high Annexin V levels seen upon incubation with 4C7 but not DHER (Fig. 1B,C). We next used flow cytometry to determine if differences in surface expression of death receptors can explain the differential TRAIL sensitivity observed. We found that all three cell lines express DR5 to a similar extend (Fig. 1D). These results reinforce the notion that death receptor expression alone is not predictive of TRAIL susceptibility.
Figure 1.
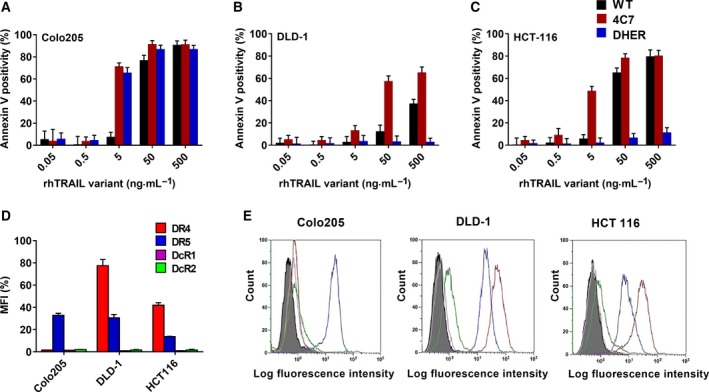
Different colon adenocarcinoma cell lines exhibit differential sensitivities via DR4 and DR5. Apoptosis inducing potential of rhTRAIL WT, 4C7 and DHER (0.05–500 ng·mL −1) in COLO 205 (A), DLD‐1 (B) and HCT 116 (C) was determined after 16 h treatment using Annexin V‐APC by flow cytometry. Cell surface expression of TRAIL receptors was determined in COLO 205, DLD‐1 and HCT 116 cells using flow cytometry analysis and depicted as the Mean Fluorescence Intensity (MFI) ratio (D) and as FACS histograms compared to the binding of isotype antibody (E). The values shown are mean ± SD of three independent experiments.
Inhibition of O‐glycosylation leads to loss of fucosylation resulting in a decrease in TRAIL sensitivity
A role for fucosylation in TRAIL‐induced apoptosis of colon adenocarcinomas has been previously implicated 24, 35, 36, 43. Wagner et al. tested a panel of 36 colorectal adenocarcinoma cell lines and found that the sensitivity to TRAIL correlated with increased mRNA levels of the O‐glycosylation initiating enzyme GALNT3, as well as the O‐glycan processing fucosyltransferase enzymes FUT3 and FUT6 24. However, they did not report the effect of inhibiting of glycosylation on the different death receptors (DR4 or DR5). Here we show that cell death induction via both DR4 and DR5 is hampered in COLO 205 upon pretreatment with the pan O‐glycosylation enzyme inhibitor benzyl 2‐acetamido‐2‐deoxy‐ α‐d‐galactopyranoside (bGalNAc) (Fig. 2A). In DR4‐sensitive HCT 116 cells, cell death induced by 4C7 was also significantly decreased after bGalNAc treatment (Fig. 2B). We have not detected changes in the level of cell death in the DR4/5‐resistant human fibroblasts or in HCT 116 cells induced with DR5‐specific rhTRAIL DHER (Fig. 2C). These results substantiate for both death receptors the importance of O‐glycosylation and successive fucosylation for becoming sensitive to TRAIL‐induced cell death.
Figure 2.
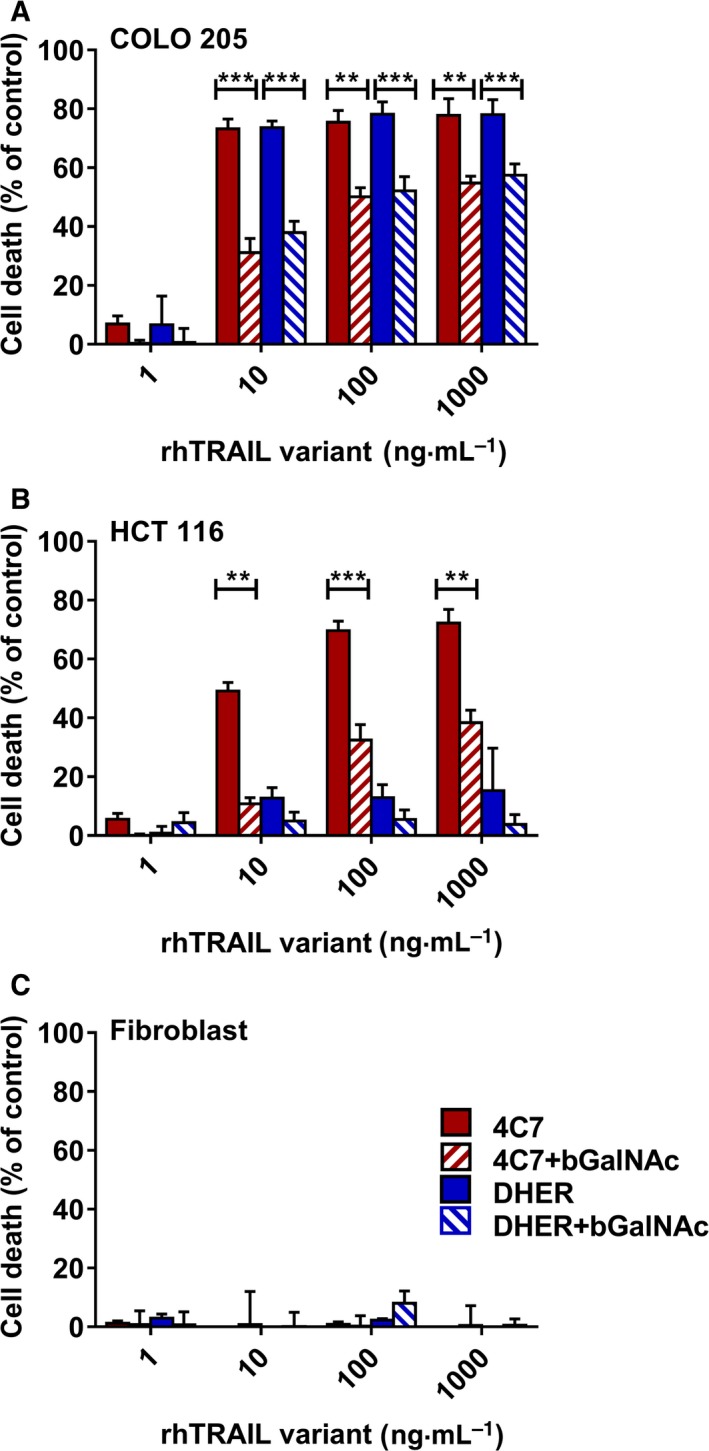
Inhibition of O‐glycosylation decreases TRAIL sensitivity. COLO 205 (A), HCT 116 (B) and fibroblasts (C) were pretreated with 2 mm bGalNAc for 24 h, after which cells were stimulated with 1–1000 ng·mL −1 rhTRAIL 4C7 or DHER for 16 h. Cell death levels were determined by MTS assay. The values shown are mean ± SD of three independent experiments. P value was analysed by one‐way ANOVA in Turkey's multiple comparison with graphpad prism version 5.00. **P < 0.005 and ***P < 0.0005.
Inhibition of fucosylation by adding 2F‐peracetyl‐fucose decreases sensitivity to DR5 specific TRAIL
In order to investigate further the effect of fucosylation on TRAIL‐induced apoptosis, we chose a fucosylation inhibitor to probe this effect in a cellular setting. Although most inhibitors only work in vitro 44, Rillahan et al. 45 showed that 2F‐peracetyl‐fucose (2FF) is cell permeable and can be used in vivo. We firstly examined the fucosylated oligosaccharides from DLD‐1 cells by blotting with recombinant biotinylated Aleuria aurantia lectin (AAL) which specifically binds to fucose 46, 47. Figure 3A shows that the fucosylation level in DLD‐1 cells reduced only after treatment with the high concentration of 2FF, which is in line with the observations from Rillahan et al. 45. Subsequently, we performed viability assays to explore alterations in the sensitivity of DLD‐1 cells to rhTRAIL WT and DHER after adding 2FF. DLD‐1 cells were firstly treated with 2FF for 3 or 5 days, followed by 24 h incubation with rhTRAIL WT or DHER. It can be seen from Fig. 3B that cell survival increased after incubating with the fucosylation inhibitor indicating that downregulation of fucosylation renders the receptor less sensitive for rhTRAIL WT and DHER. This decreased cell death induced by rhTRAIL DHER was further confirmed by flow cytometric analysis detecting apoptosis using the Violet Ratiometric Membrane Asymmetry Probe. Figure 3C shows that frequency of early apoptotic cells decreased after pretreatment with 500 μm of 2FF, which means that low fucosylation levels lead to a reduction in rhTRAIL DHER‐induced apoptosis.
Figure 3.
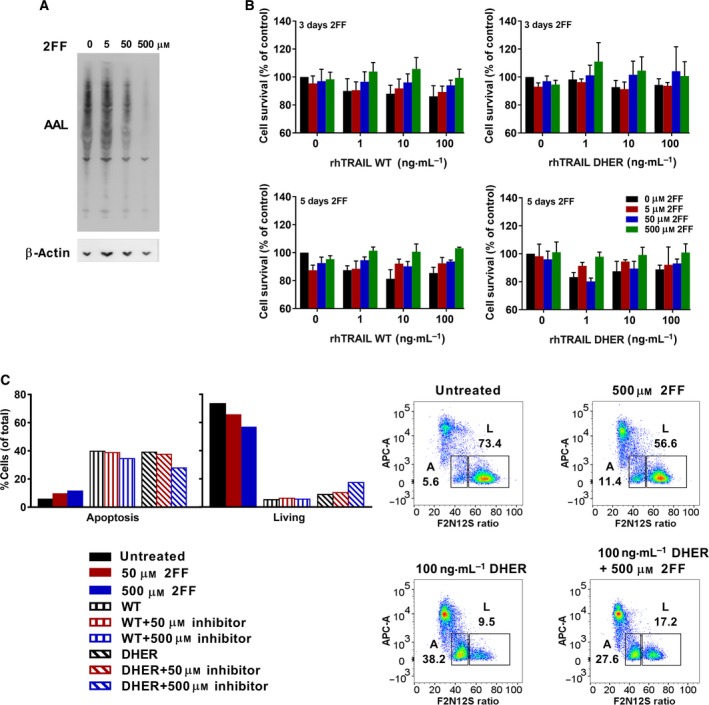
Inhibition of fucosylation by adding 2F‐peracetyl‐fucose decreases sensitivity to DR5 specific TRAIL. (A) Proteins were blotted by recombinant biotinylated AAL and probed again by β‐actin as loading control. (B) Cell survival was measured and analysed by MTS assay. The values shown are mean ± SD of three independent experiments. (C) Cells were incubated with 2FF for 3 days and followed by adding 100 ng·mL −1 rhTRAIL DHER for 24 h. Apoptotic cells were stained by Violet Ratiometric Membrane Asymmetry Probe and measured by LSR‐II. L stands for living cells, A stands for apoptotic cells.
Triggering the fucosylation salvage pathway by l‐fucose improves TRAIL sensitivity
Fucosylation is an important type of posttranslational modification in colon cancer 31 and fucosyltransferase enzyme levels are important determinants. However, the presence of the donor substrate GDP‐fucose is equally important. Therefore, we reasoned that targeting the de novo pathway or inducing the salvage pathway by increasing the level of GDP‐fucose donor might enhance fucosylation and subsequently trigger TRAIL‐mediated apoptosis. Recently, Moriwaki et al. 35 showed that HCT 116 cells are less able to synthesize GDP‐fucose, due to mutations in the GMDS gene that plays a critical role in the de novo GDP‐fucose pathway. We therefore investigated whether the salvage pathway can be activated by the addition of l‐fucose, and thereby potentially sensitize these cells to TRAIL‐induced cell death. To do this, COLO 205, DLD‐1, HCT 116, SW948 and WiDr cells were preincubated with l‐fucose and after 24 h further treated with rhTRAIL WT, 4C7 or DHER for 16 h (0.05–500 ng·mL−1). The results show that the induction of DR5‐mediated cell death is clearly enhanced upon pretreatment with l‐fucose in DR5 resistant cell lines DLD‐1, HCT 116, SW948 and WiDr (Fig. 4B–E), with rhTRAIL DHER activities going up between 5 and 10 times in the presence of l‐fucose. In contrast to DHER sensitive COLO 205 cells, we only detected a slight enhancement of TRAIL‐induced cell death using rhTRAIL DHER at low concentration (0.05 ng·mL−1) after l‐fucose pretreatment (Fig. 4A). Since COLO 205, DLD‐1 and HCT 116 are already quite sensitive to 4C7, it is conceivable that it is difficult to increase the cell death at relative high concentration (50 or 500 ng·mL−1). However, enhanced cell death was observed in COLO 205 cells after treatment with low concentration 4C7 (Fig. 4A). The response to rhTRAIL WT was increased to an intermediate level in the presence of l‐fucose reflecting its signalling via both DR4 and DR5. Enhanced cell death was not due to changed expression levels of DR4 and/or DR5 receptors (Fig. 5A), neither due to the treatment by l‐fucose alone (Fig. 5B).
Figure 4.
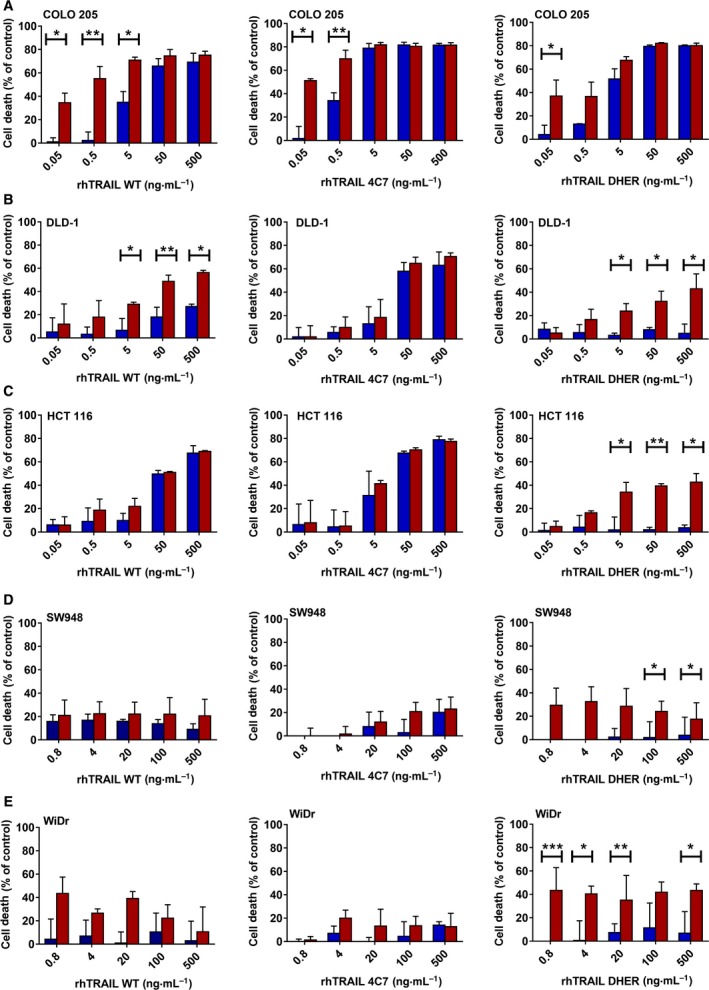
l‐fucose treatment augments TRAIL‐induced apoptosis predominantly via the activation of DR5. COLO 205 (A), DLD‐1 (B), HCT 116 (C), SW948 (D) and WiDr (E) cells were pretreated with 0 or 50 mm l‐fucose for 24 h and subsequently incubated with 0.05–500 ng·mL −1 rhTRAIL WT, DHER or 4C7 for another 16 h. Cell death was assessed using MTS assay. Blue bars represent cells without adding l‐fucose and red bars represent cells pretreatment with 50 mm l‐fucose. The values shown are mean ± SD of three independent experiments. P value was analysed by one‐way ANOVA in Turkey's multiple comparison with graphpad prism version 5.00. *P < 0.05, **P < 0.005 and ***P < 0.0005.
Figure 5.
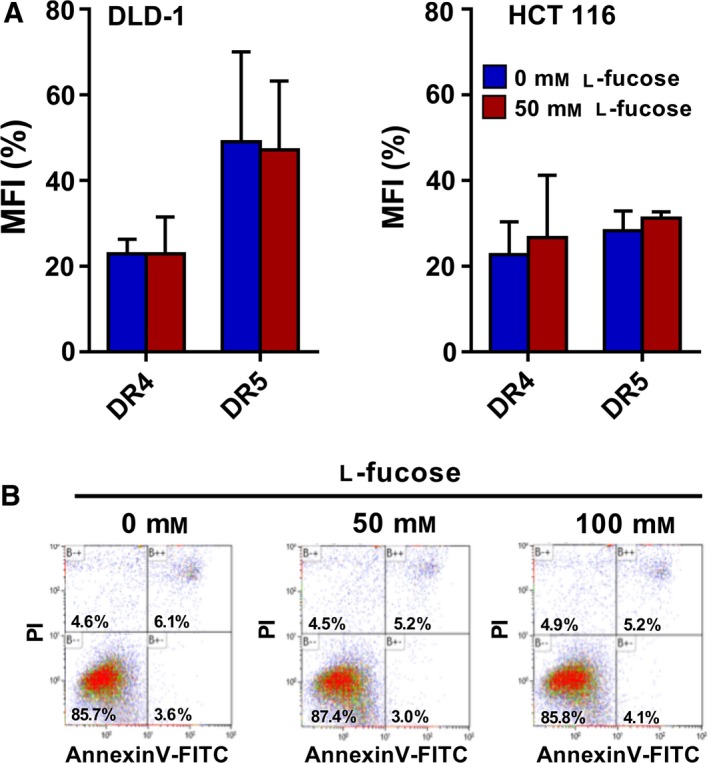
Death receptor 4 and DR5 receptor expression and l‐fucose tolerance. (A) Cell surface expression of TRAIL receptors was determined in DLD‐1 and HCT 116 cells treated with 0 or 50 mm l‐fucose using flow cytometry analysis and expressed as the Mean Fluorescence Intensity (MFI) ratio compared to the binding of isotype antibody. The values shown are mean ± SD of three independent experiments. (B) HCT 116 cells were treated with 0, 50 or 100 mm l‐fucose and cell death was measured by FACS using Annexin V‐FITC/PI staining.
Taken together, these results indicate that l‐fucose sensitizes DLD‐1, HCT 116, SW948 and WiDr to cell death through the activation of mainly DR5.
Stable and transient overexpression of FUT3 and FUT6 enhances TRAIL sensitivity via both death receptors
Our previous data show that upregulation of fucosylation by the salvage pathway indeed improves the sensitivity to TRAIL. Next, we focused on the two specific fucosyltransferase enzymes FUT3 and FUT6, which associate to TRAIL‐sensitivity in colon cancer cells as reported by Wagner et al. Quantitative PCR analysis confirmed the low mRNA levels of FUT3 and FUT6 in our DR5‐insensitive DLD‐1 and HCT 116 cells (Fig. 6A), which correlates with previous observations 24. We generated stable cell lines expressing either FUT3 or FUT6 in both DLD‐1 and HCT 116. Cells were transduced either with an empty lentiviral vector (CTRL) or a vector expressing FUT3 or FUT6 with a GFP gene, allowing for the selection of transduced cells. Using qRT‐PCR a clear increase in relative FUT3 or FUT6 mRNA levels was observed compared to control with DLD‐1 cells expressing higher levels of FUT3 and FUT6 compared to HCT 116 (Fig. 6A). Additional western blot analysis of FUT3 expression, showed a significant increase in FUT3 protein levels in both DLD‐1:FUT3 and HCT 116:FUT3 cells, when compared to control and FUT6 overexpressing cells (Fig. 6B). FUT6 protein levels could not be analysed with western blot due to the lack of suitable antibodies. Flow cytometry analysis of transduced DLD‐1 and HCT 116 cells showed that overexpression of FUT3 and FUT6 did slightly change the expression of DR4 and DR5, but not to a level that explains the change in sensitivity (Fig. 6C).
Figure 6.
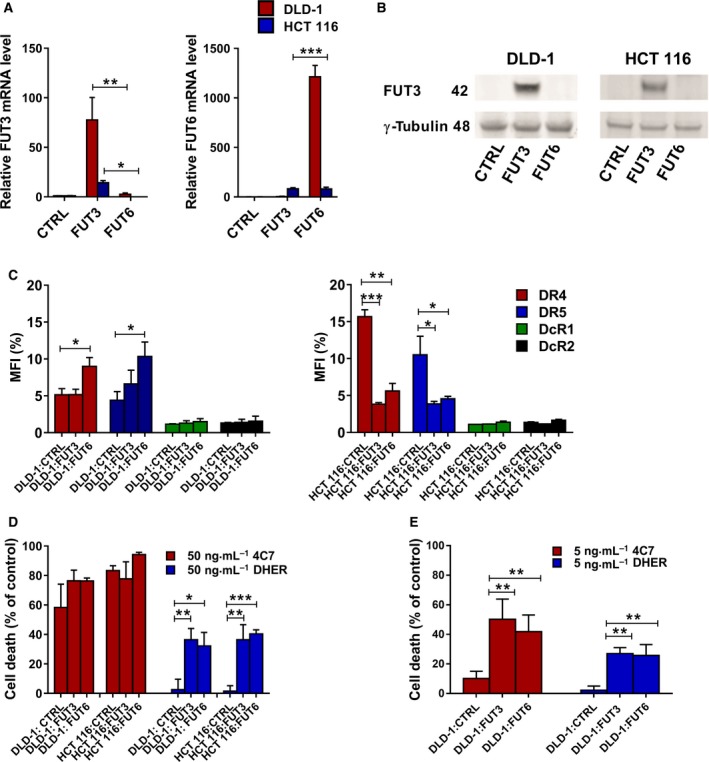
Fucosyltransferase 3 and FUT6 overexpression enhances TRAIL sensitivity of DLD‐1 and HCT 116 cells via both death receptors. Overexpression of FUT3 and FUT6 was analysed by qRT‐PCR of DLD‐1 and HCT 116 cells transduced with control, FUT3 or FUT6 overexpressing plasmid. (A) The amount of FUT3 or FUT6 amplicon was relative to the endogenous reference RPL27 and normalized to the control cells. (B) Western blot analysis of FUT3 expression in control or FUT3/6 transduced DLD‐1 and HCT 116 cells, γ‐tubulin served as a loading control. (C) The expression of DR4, DR5, DcR1 and DcR2 in control, FUT3 and FUT6 overexpressing DLD‐1 and HCT 116 cells. Cell death of transduced DLD‐1 and HCT 116 cells overexpressing FUT3/6 was assessed after treatment with 50 ng·mL−1 rhTRAIL 4C7 or DHER (D) or 5 ng·mL−1 rhTRAIL 4C7 or DHER (E) for 16 h as measured by MTS assay. The values shown are mean ± SD of three independent experiments. P value was analysed by one‐way ANOVA in Turkey's multiple comparison with graphpad prism version 5.00.*P < 0.05, **P < 0.005 and ***P < 0.0005.
To evaluate the impact of FUT3 or FUT6 stable overexpression on TRAIL‐mediated apoptosis via either DR4 or DR5, the transduced cells were treated with 50 ng·mL−1 of the 4C7 or DHER, respectively. Overexpression of either FUT3 or FUT6 dramatically enhanced TRAIL‐induced cell death via DR5 in both DLD‐1 and HCT 116 cells (Fig. 6D). The sensitivity to rhTRAIL 4C7 in FUT3 or FUT6 overexpressing DLD‐1 cells only slightly improved, whereas cell death remained largely unaltered in overexpressing HCT 116 cells (Fig. 6D). This is mainly due to the already high cell death in control cells using 50 ng·mL−1 rhTRAIL 4C7. Interestingly at low concentrations (5 ng·mL−1), where control DLD‐1 cells show ~ 10% of killing with rhTRAIL 4C7, transduced cells show an increase in killing cells, indicating that only at lower concentrations DR4‐mediated cell death is slightly improved by the overexpression of fucosyltransferases (Fig. 6E). Moreover, to rule out the possibility that random integration of the gene into the genome itself could cause the differential TRAIL sensitivity we repeated the sensitivities assays with transient FUT3 and FUT6 overexpressing DLD‐1 cells. Overexpressing FUT3 show a similar increase in TRAIL‐induced cell death, which indicates that the observations are robust (Fig. 7A). Transient overexpression of FUT6 in both DLD‐1 and HCT 116 cells also enhanced the DR5 mediated cell death as seen by Annexin V‐APC staining (Fig. 7B). In conclusion, FUT3 and FUT6 have a role in the regulation of TRAIL sensitivity via both death receptors and are most important for DR5.
Figure 7.
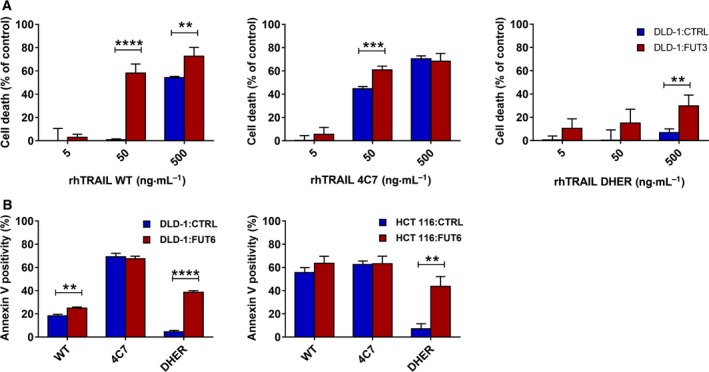
TRAIL sensitivity of transient FUT3 or FUT6 transfected DLD‐1 and HCT 116 cells. Transient FUT3 overexpressing DLD‐1 cells were treated with 5–500 ng·mL−1 rhTRAIL WT, 4C7 or DHER and cell death was measured by MTS assay (A). (B) Transient FUT6 overexpressing cells lines DLD‐1 and HCT 116 cells were treated with 50 ng·mL−1 rhTRAIL WT, 4C7 and DHER. Apoptosis induction was assessed after 16 h using Annexin V‐APC staining measured by flow cytometry. The values shown are mean ± SD of three independent experiments. P value was analysed by one‐way ANOVA in Turkey's multiple comparison with graphpad prism version 5.00. **P < 0.005, ***P < 0.0005 and ****P < 0.00005.
FUT3 and FUT6 overexpression leads to preclustering of DR4 and DR5
Caspase‐8 is very important in apoptotic signal transduction via ligand‐dependent receptor activation. For TRAIL‐mediated apoptosis, the signal is transmitted into the pathway by the formation of the DISC, which further recruits and activates caspase‐8 48. To investigate whether the observed higher levels of apoptosis in FUT3 overexpressing cells were mirrored in the DISC formation, we co‐immunoprecipitated the DISC proteins using FLAG‐rhTRAIL WT (FLAG‐WT) and FLAG‐DHER, respectively. Caspase‐8 activation was shown to be more pronounced in the DISC in control and FUT3 overexpression cells when immunoprecipitated with FLAG‐WT and less by FLAG‐DHER (Fig. 8A). This is in line with the binding properties of FLAG‐WT, which can trigger the DISC formation via both DR4 and DR5, while FLAG‐DHER can do this only via DR5. Notably, more DISC‐associated caspase‐8 was found in the S and T clonal populations of the FUT3 overexpressing cells as compared to the control cells, indicating that the level of apoptotic signalling activation in the FUT3 overexpressing cells is higher. We also show that DR5 participating in the DISC formation is more noticeable in the immunoprecipitates of FUT3 overexpressing cells, reinforcing the crucial response of DR5 to fucosylation (Fig. 8A). In a screen across a panel of breast cancer cell lines, TRAIL‐sensitive cells showed more caspase‐8 activity than TRAIL resistant cells 49. This has also been found in a subset of colorectal cancer cell lines 24. In order to examine caspase‐8 and the downstream signal in more detail, we analysed the pro‐caspase‐8, cleaved caspase‐8 and downstream PARP‐1 activation. Western blot analysis demonstrated that activation and cleavage of caspase‐8 and PARP‐1 was more pronounced in FUT3 and FUT6 overexpressing cells compared to control, which confirms that the effect of fucosylation is transmitted downstream in the apoptosis pathway (Fig. 8B). Moreover, the amount of cleaved caspase‐8 and PARP‐1 increased after the treatment with 4C7 and DHER, which indicated that this upregulation of fucosylation improved the DISC formation via both death receptors. We therefore investigated whether fucosylation enhanced TRAIL sensitivity by preclustering of DR4 and DR5 on the cell surface using immunofluorescent staining. Indeed, FUT3 and/or FUT6 overexpressing cells showed more preclustering of DR4 and DR5 compared to control cells (Fig. 8C). These results confirm that fucosylation by FUT3 and FUT6 overexpression enhances TRAIL‐induced apoptosis through DISC formation by triggering the preclustering of DR4 and DR5.
Figure 8.
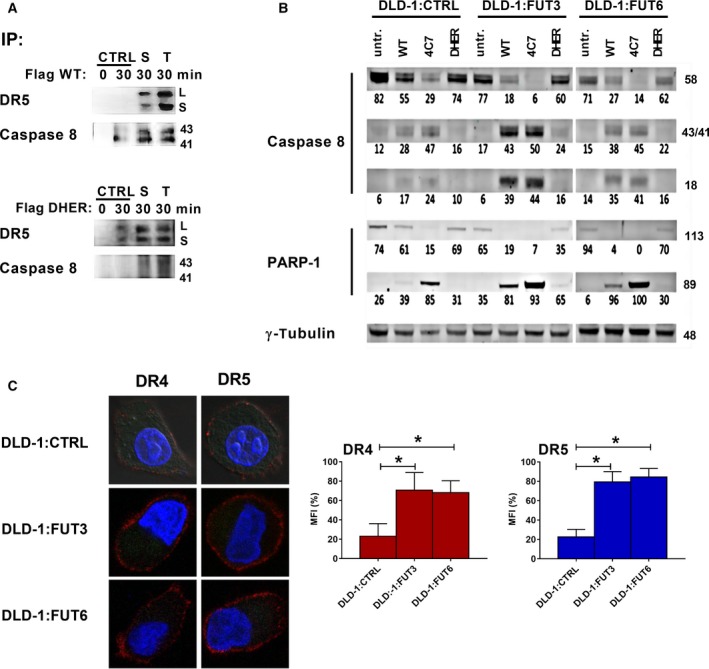
Fucosyltransferase 3 and FUT6 overexpression leads to preclustering of DR4 and DR5. (A) Co‐immunoprecipitation of the TRAIL DISC. Two population of FUT3 overexpression cells (S and T) and control cells were stimulated by 1 μg·mL −1 FLAG‐TRAIL WT or FLAG‐DHER respectively for 0 or 30 min. Then the DISC was immunoprecipitated with FLAG antibody. DR5 or cleaved caspase‐8 from the DISC were detected by immunoblot. (B) Western blot analysis of FUT3 or FUT6 transduced DLD‐1 cells treated with 500 ng·mL−1 rhTRAIL WT, 4C7 or DHER for 1 h. Caspase‐8 and PARP‐1 activation was examined and analysed using densitometry (values depicted as % of total protein); γ‐tubulin served as a loading control. The data are presented as mean values of three independent experiments. (C) Immunostaining of DLD‐1 cells transduced with control, FUT3 or FUT6 plasmids, containing the GFP gene. Cells were seeded on poly‐L‐lysine coated coverslips and stained with primary antibodies against DR4 or DR5, and secondary antibody conjugated with Alexa Fluor 647. Nuclei were counterstained with DAPI. The right graphs depict the quantification of the mean fluorescence intensity of 20 single confocal pictures of each cell lines. P value was analysed by one‐way ANOVA in Turkey's multiple comparison with graphpad prism version 5.00. *P < 0.05.
Discussion
Recent results have shown that glycosylation of death receptors plays an important role in TRAIL‐mediated apoptosis. In particular, it was shown that O‐glycosylation increased TRAIL sensitivity by inducing efficient DISC formation and caspase‐8 activation through the clustering of death receptors 24. Moreover, the N‐glycosylation via human DR4 and mouse death receptor also regulates TRAIL‐induced cell death by the enhancing DISC formation 37. The fact that O‐ and N‐glycosylation can undergo successive posttranslational modification, such as fucosylation and sialylation, implies that there might be a connection and an additive effect in relation to TRAIL sensitivity 33, 50. Specifically, FUT3 and FUT6 expression was shown to correlate with TRAIL sensitivity in a large panel of colorectal adenocarcinoma cell lines 24. Moreover, GMDS deficiency leading to a loss of de novo fucosylation was found to increase tumour growth and metastasis in vivo 35, due to a reduced FADD‐dependent complex II formation 43. Here, we provide for the first time evidence that signalling via DR5 is for the major part responsible for this l‐fucose dependent apoptosis induction.
In this study, we observed that DLD‐1 and HCT 116 cells are highly insensitive to DR5‐mediated apoptosis in contrast to the colon adenocarcinoma COLO 205 cell line that shows a high sensitivity to both DR4‐ and DR5‐mediated apoptosis. Here we further show that this insensitivity is not related to a lack of surface expression levels of DR5, but to a specific lack of fucosylation. This finding is in line with earlier observations that TRAIL‐sensitive colon adenocarcinoma cell lines show relatively high mRNA levels of FUT3 and FUT6, while the TRAIL‐resistant cell lines, such as DLD‐1 and HCT 116, show low expression of FUT3 and FUT6 24, 37. Reduction in the number of fucosyl attachment sites by pretreatment with the O‐glycosylation inhibitor bGalNAc led to decreased TRAIL sensitivity via DR5 in all three cell lines mentioned, as determined by us using TRAIL‐death receptor specific variants. The bGalNAc treatment also had an effect on the DR4 sensitivity in HCT 116, although there is a considerable residual activity. This may be due to the fact that DR4, in contrast to DR5, has N‐glycosylation sites, which are not affected by bGalNAc ensuing outstanding attachment sites for fucose. This observed reduction in DR4 signalling is slightly different from the data reported by Dufour et al., in which they reported that DR4‐induced apoptosis in another bone osteosarcoma cell line was not at all affected by the bGalNAc treatment. This may be due to the usage of a different cell line and/or the use of a different DR4‐specific variant 37. Direct inhibition of fucosylation turned out to be complicated as most reported inhibitors do not penetrate into the cell and can be used only in vitro. By exploiting promiscuous monosaccharide salvage pathways the inhibitor 2FF can be partly imported and metabolized in cells 45 and we indeed could show inhibition of fucosylation in DLD‐1 colon cancer cells using high concentrations. The observation that the sensitivity to rhTRAIL DHER in these cells is lowered, forms an additional indication for the important role of fucosylation in activating death receptor.
Without doubt, the most spectacular finding of our study is the observation that the simple addition of l‐fucose re‐sensitizes DLD‐1 and HCT 116 cells to DR5‐mediated apoptosis and enhances the DHER sensitivity from 5 to 40%. Interestingly, oral administration of l‐fucose was found to successfully increase fucosylation in the patient with leukocyte adhesion deficiency type II (LAD II), a rare inherited disorder of fucose metabolism 51, 52, 53. Moreover, tumour growth, mitotic activity and metastatic setting were greatly suppressed by daily intraperitoneal injection of l‐fucose in the Ehrlich carcinoma mice 53. Our results further suggest that the sensitivity to TRAIL can be simulated after elevated expression of FUT3 or FUT6 in tumour cells, which is mainly mediated via DR5 as evidenced by the enhanced sensitivity to DHER of these cells.
Furthermore, mechanistic studies show that in FUT3/6 overexpressing cells compared to control cells, the preclustering of DR4 and/or DR5 is enhanced without any increase in the level of receptor expression. Moreover this clustering directly stimulates more DISC formation and caspase‐8 activation. In agreement with the regulation by O‐ or N‐glycosylated death receptors, the upregulation of fucosylation changes the distribution of death receptors and enhances their clustering leading to enhanced apoptotic signal transduction after the stimulation by TRAIL 24, 39. There is some evidence showing that a preligand assembly domain association (PLAD) consisting of death receptors, especially DR5, and decoy receptors lacking a death domain attenuates TRAIL‐induced apoptosis 14, 54. In concert with this PLAD model, it can be conceived that if most death receptors are involved in the preclustering activity due to the upregulation of fucosylation, the possibility for them to bind to the decoy receptors greatly decreases. By this mechanism, TRAIL induced apoptosis might become significantly improved upon fucosylation.
Many examples have already shown the diagnostic applications related to fucosylation. The most representative type of cancer biomarker is fucosylated alpha‐fetoprotein (AFP) in hepatocellular carcinoma (HCC). AFP with core fucosylation is very specific in the early stage of HCC and widely used as diagnostic marker 55, 56. Kyselova et al. 57 measured the glycan profile in serum and proposed eight potential sialylated and fucosylated N‐glycan structures to stage the progression of breast cancer. Moreover, in the serum from patients with advanced pancreatic cancer, haptoglobin fucosylation patterns were found that differ from those at early stage 58. Our results suggest that the status of fucosylation indicates the sensitivity to DHER in colon carcinoma and provides a potential biomarker for TRAIL therapy. Taken together, our findings give a new insight into the effect of posttranslational modification on TRAIL‐sensitivity and imply promising novel approaches for restoring TRAIL response in resistant cancer cells.
Materials and methods
Cell lines and reagents
Human colorectal cancer cell lines COLO 205, DLD‐1, HCT 116, SW948 and WiDr were cultured in RPMI1640 medium supplemented with 10% fetal bovine serum (FBS), 100 units·mL−1 Penicillin and 100 μg·mL−1 Streptomycin in a humidified incubator at 37 °C containing 5% CO2. Human foreskin fibroblasts (Cell Stock, University of Groningen) were cultured in Ham's F‐10 medium supplemented with 10% fetal bovine serum (FBS), 100 units·mL−1 Penicillin and 100 μg·mL−1 Streptomycin. All materials mentioned above were obtained from Thermofisher Scientific (Landsmeer, The Netherlands). rhTRAIL wild‐type (WT), the DR4‐specific TRAIL variant 4C7 and the DR5‐specific TRAIL variant DHER (amino acids 114–281) were constructed and produced as previously described 37, 38. l‐(‐)‐Fucose (l‐fucose), benzyl 2‐acetamido‐2‐deoxy‐α‐d‐galactopyranoside (bGalNAc) and 2F‐peracetyl‐fucose (2FF) were purchased from Merck (Darmstadt, Germany).
MTS assay
Cell viability and proliferation assays were conducted using MTS assays. Cells were seeded in triplicate in 96‐well plates at a cell density of 10 000 cells/well in 0.1 mL medium. After 24 h, cells were treated with concentrations ranging from 0 to 500 ng·mL−1 of rhTRAIL WT, 4C7 or DHER for 16 h, to a final volume of 0.15 mL. In the case of only l‐fucose treatment, cells were incubated with different concentrations of l‐fucose (0–100 mm) for 48 h to a final volume of 0.2 mL. For the combination of l‐fucose and rhTRAIL, cells were preincubated with 0 or 50 mm of l‐fucose in a final volume of 0.15 mL. After 24 h, cells were treated with concentrations ranging from 0 to 500 ng·mL−1 of rhTRAIL WT, 4C7 or DHER for 16 h to a final volume of 0.2 mL. Stock solutions of l‐fucose were serially diluted in serum‐free RPMI medium and TRAIL ligands in complete medium. Cells were incubated with MTS reagent according to manufacturer's instructions (Promega, Leiden, The Netherlands). Cell viability was determined by measuring the absorption at 492 nm using a microplate reader (Thermo Labsystems, Vasai, India). For inhibition of O‐glycosylation, cells were pretreated with 2 mm bGalNAc in a final volume of 0.15 mL. After 24 h, cells were treated with rhTRAIL concentrations, ranging from 0 to 1000 ng·mL−1 of rhTRAIL 4C7 or DHER for 16 h, to a final volume of 0.2 mL. For inhibition of fucosylation, cells were seeded at the density of 1000 cells/well or 100 cells/well in 0.1 mL medium and incubated with 2FF at indicated concentration for 3 days and 5 days, respectively. Next, cells were treated with rhTRAIL WT or DHER at indicated concentration for 24 h.
Apoptotic assay
Apoptosis induction was measured using Annexin V‐APC (IQP Products, Groningen, The Netherlands) staining and quantified by flow cytometry. Cells were seeded in 6‐well plates 24 h prior to treatment. The next day, cells were incubated with 0–500 ng·mL−1 rhTRAIL WT, 4C7 or DHER for 16 h. After treatment, cells were harvested and washed with calcium buffer (10.9 μm HEPES, 140 μm NaCl, 2.5 μm CaCl2) (Merck). Cell pellets were resuspended in 60 μL calcium buffer complemented with 5 μL Annexin V‐APC and incubated for 20 min on ice. Cells were washed and analysed using a FACS Calibur flow cytometer (BD Biosciences). For cells treated by 2FF, 3E5 cells were firstly seeded in one‐well of 6‐well plates. Fifty micrometres or 500 μm 2FF was added at the following day and incubated for 3 days. Three days later, 100 ng·mL−1 rhTRAIL WT or DHER was added for 24 h. After treatment, cells were harvested in PBS and apoptotic cells were measured and analysed by LSR‐II (BD Biosciences) using the dead cell apoptotic kit from Thermofisher Scientific (Violet Ratiometric Membrane Asymmetry Probe, Landsmeer, the Netherlands).
TRAIL receptor membrane expression analysis
Cells were harvested and washed with FACS buffer (PBS with 1% BSA). Cell surface expression of TRAIL receptors was determined using 10 μg·mL−1 TRAIL‐R1, TRAIL‐R2, TRAIL‐R3, TRAIL‐R4 (Alexis Biochemicals, Enzo Life Sciences, Bruxelles, Belgium) or negative control mouse IgG1 (Agilent, Santa Clara, CA, USA). Cells were incubated with primary antibodies for 1 h on ice. Subsequently, the cells were washed and incubated for 1 h with R‐phycoerythrin (PE) conjugated goat anti‐mouse antibody (Southern Biotech, Birmingham, AL, USA) or Alexa Fluor 488 conjugated goat anti‐mouse antibody (Thermofisher Scientific). Receptor cell surface expression was analysed using a FACS Calibur flow cytometer (BD Biosciences).
Transient overexpression of FUT6‐GFP and lentiviral overexpression of FUT3 and FUT6
For transient overexpression of FUT6‐GFP, DLD‐1 and HCT 116 cells were seeded in 6‐well plates at a density of 150 000 cells/well. The next day, the subconfluent cultures were transfected with plasmid containing GFP‐conjugated α‐1,3‐fucosyltransferase 6, fuc‐T6‐GFP (FUT6‐GFP) (a kind gift from prof. Jack Rohrer) using the FuGENE HD Transfection Reagent (Promega, Leiden, the Netherlands) according to manufacturer's instructions. Cells were seeded 48 h after transfection and treated with 50 ng·mL−1 rhTRAIL WT, 4C7 or DHER for 16 h. Cell death induction was measured using Annexin V‐APC staining.
For lentiviral overexpression of FUT3 and FUT6, pLenti‐GIII‐CMV‐GFP‐2A‐Puro plasmids encoding FUT3 or FUT6 and the control plasmid were purchased from Applied Biological Materials Inc. (Heidelberg, Germany). For the packaging of the lentiviral particles, 2*106 HEK293 cells were plated in 94 mm cell culture dishes. The following day, cells were transfected with either pLenti‐CMV‐GFP vector expressing FUT3, FUT6 or the control vector, using CaCl2. After 48 h, the medium containing virus particles was harvested, filtered, mixed with Polybrene (Merck) and added to DLD‐1 and HCT 116 cells, which were plated the day before at a density of 0.25*106 cells in a 6‐well plates; the final concentration of Polybrene was 10 μg·mL−1. The following day, the previous steps were repeated, therefore cells were exposed to the viral particles for 48 h in total. Mixed populations of control‐, FUT3‐ or FUT6‐overexpressing cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 units·mL−1 Penicillin, 100 μg·mL−1 Streptomycin and 2 μg·mL−1 Puromycin (Merck) in a humidified incubator at 37 °C containing 5% CO2. Single clones that expressed GFP were subcloned.
Quantitative real‐time PCR
RNA was isolated from DLD‐1 and HCT 116 cells transduced with control, FUT3 or FUT6 lentiviral plasmids using the RNeasy Mini Kit (QIAGEN, Venlo, the Netherlands) according to the manufacturer's instructions. cDNA was synthesized from 4 μg total RNA using oligo dT primers and M‐MLV Reverse Transcriptase (Thermofisher Scientific) in a total volume of 80 μL. Quantitative real‐time (qRT)‐PCR was performed to determine the mRNA expression levels of FUT3 (5′‐ GGACATGGCCTTTCCAC ATC‐3′ and 5′‐TCCAGGTGCTGGCAGTTAGG‐3′), FU T6 (5′‐CGCTTCCCAGACAGCACAGG‐3′ and 5′‐TCCGTCCATGGCTTTCAGCTGCCA‐3′) and the housekeeping gene RPL27 (5′‐TCCGGACGCAAAGCTGTCATCG‐3′ and 5′‐TCTTGCCCATGGCAGCTGTCAC‐3′) using SsoAdvanced Universal SYBR Green Supermix (BioRad, Veenendaal, The Netherlands) on the CFX Connect Real‐Time PCR Detection System (Bio‐Rad). The protocol was as follows: initial denaturation at 98 °C for 3 min, followed by 45 cycles of amplification (5 s at 98 °C and 20 s at 65 °C). Finally, a melting curve analysis was performed to ensure that only a single PCR amplicon was produced.
Western blotting
Cells were harvested and lysed by using the Mammalian Protein Extraction Reagent (Thermofisher Scientific) with additional Protease Inhibitor Cocktail, EDTA‐Free (Roche, Basel, Switzerland). Protein concentrations were determined using a Bradford assay (Bio‐Rad). Equal amounts of protein (20 μg) for each sample were loaded per lane on precast 4–12% SDS‐PAGE gels (Thermofisher Scientific) and transferred onto Immobilon‐FL PVDF 0.45 μm membranes (Merck). Subsequently, the membranes were blocked for 1 h at room temperature in blocking buffer (Rockland, Limerick, USA). Western blot membranes were probed overnight at 4 °C. The following primary antibodies were used: Caspase‐8 (Cell Signalling Technologies, Leiden, the Netherlands), FUT3 (Abcam, Cambridge, UK) and PARP‐1 (Cell Signalling Technologies). Goat‐α‐mouse‐IRDye 680 (LI‐COR Biosciences, Lincoln, NE, USA) or goat‐α‐rabbit‐IRDye 800CW (LI‐COR Biosciences) secondary antibodies were used for detection using a LI‐COR Odyssey Infrared Imaging System (Badhomburg, Germany). Membranes were probed with anti‐γ‐tubulin (Merck) to confirm equal loading. For AAL blot, after proteins were transferred onto nitrocellulose membrane (Merck), this membrane was blocked overnight in 3% BSA at 4 °C. Afterwards, membrane was incubated with 0.5 μg·mL−1 of recombinant biotinylated AAL (Vector lab, Peterborough, UK) for 30 min at room temperature. Streptavidin conjugated with HRP (Thermofisher Scientific) used as secondary antibody was added for 30 min at room temperature. At last, membrane was detected using Pierce ECL kit (Thermofisher Scientific). Anti‐β‐actin (Cell Signalling Technology) was probed as relative loading control.
Co‐immunoprecipitation of TRAIL‐DISC complex
M‐270 Epoxy beads (Thermofisher Scientific) were covalently coupled with anti‐FLAG antibody (Merck) according to the manufacture's instruction (5–7 μg per mg beads) at 37 °C for 20 h. The next day beads were washed and stored following the protocol from the company. 3–4 × 107 cells were harvested and incubated with 1 μg·mL−1 FLAG‐rhTRAIL or FLAG‐DHER in the complete medium at 4 °C to prevent ligand receptor complex internalization. After washing with PBS the cell pellet was weighted and nine volumes of the extracting buffer A (Thermofisher Scientific) supplemented with 50 mm NaCl and protease inhibitor cocktail, were added and incubated on ice for 15 min. Cell lysate was cleared by centrifugation at 2600 g for 5 min at 4 °C, then the DISC was co‐immunoprecipitated overnight with the prepared beads at 4 °C. Beads were washed three times with the extraction buffer A and one time with 1xLWB supplied with the kit; then the protein complex was eluted using SDS loading buffer (50 mm Tris‐HCl, pH 6.8, 2% SDS, 6% glycerol).
Immunostaining of surface DR4 and DR5
DLD‐1:CTRL, DLD‐1:FUT3 and DLD‐1:FUT6 cells were seeded at a density of 150 000 cells on Poly‐l‐lysine (Merck) coated coverslips. Cells were fixed using 4% formaldehyde solution (Merck) for 15 min at room temperature. The cells were then stained for 1 h with 1 : 50 in PBS diluted TRAIL‐R1 (Alexis Biochemicals, Enzo Life Sciences, Bruxelles, Belgium), DR5‐01‐1 (Exbio, Praha, Czech republic) or IgG1 negative control (Agilent). After washing with PBS three times, cells were incubated with secondary antibody donkey anti‐mouse IgG (H + L) Alexa Fluor 647 (Jackson ImmunoResearch, Cambridge, UK) at a concentration of 1 : 100 for 1 h. Nuclei were counterstained with 0.2 μg·mL−1 DAPI (Thermofisher Scientific) for 10 min. The coverslips were mounted with CitiFluor (Agar Scientific, Stansted, UK). Slides were photographed using a Leica DMI 6000 inverted microscope.
Statistical analysis
Data were presented as mean ± standard deviation (SD) from three independent experiments. Comparisons between groups were analysed by one‐way ANOVA with graphpad prism version 5.00 (GraphPad Software, San Diego, CA, USA). Results were considered statistically significant at 5% level.
Conflicts of interest
There are no conflicts of interest.
Author contributions
BZ, IR, CR, RS and WQ designed the project. BZ, IR, CR, RS performed the experiments. BZ, IR, CR, RS and WQ analysed the results and wrote the paper.
Acknowledgements
This research was partly funded by The Dutch Technology Foundation (STW) (grant 11056), European Fund for Regional Development (KOP/EFRO) (grants 068 and 073) and the Ubbo Emmius Foundation of the University of Groningen. Part of the work has been performed at the UMCG Imaging and Microscopy Centre (UMIC). Financial support from the program of China Scholarship Council (CSC) during the PhD of Baojie Zhang.
Baojie Zhang and Ingrid A.M. van Roosmalen contributed equally to this work
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA & Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68, 394–424. [DOI] [PubMed] [Google Scholar]
- 2. Brian MW & Mayer RJ (2009) Systemic treatment of colorectal cancer. Gastroenterology 134, 1296–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chadi SA, Malcomson L, Ensor J, Riley RD, Vaccaro CA, Rossi GL, Daniels IR, Smart NJ, Osborne ME, Beets GL et al (2018) Factors affecting local regrowth after watch and wait for patients with a clinical complete response following chemoradiotherapy in rectal cancer (InterCoRe consortium): an individual participant data meta‐analysis. Lancet Gastroenterol Hepatol 1253, 1–12. [DOI] [PubMed] [Google Scholar]
- 4. Puig I, López‐Cerón M, Arnau A, Rosiñol Ò, Cuatrecasas M, Herreros‐de‐Tejada A, Ferrández Á, Serra‐Burriel M, Nogales Ó, Vida F et al (2019) Accuracy of the narrow‐band imaging International Colorectal Endoscopic Classification System in Identification of Deep Invasion in Colorectal Polyps. Gastroenterology 156, 75–87. [DOI] [PubMed] [Google Scholar]
- 5. Hall MA & Cleveland JL (2007) Clearing the TRAIL for Cancer Therapy. Cancer Cell 12, 4–6. [DOI] [PubMed] [Google Scholar]
- 6. Ashkenazi A, Pai RC, Fong S, Leung S, Lawrence DA, Marsters SA, Blackie C, Chang L, Mcmurtrey AE, Hebert A et al (1999) Safety and antitumor activity of recombinant soluble Apo2 ligand. J Clin Invest 104, 155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lawrence D, Shahrokh Z, Marsters S, Achilles K, Shih D, Mounho B, Hillan K, Totpal K, Deforge L, Schow P et al (2001) Differential hepatocyte toxicity of recombinant Apo2L/TRAIL versions. Nat Med 364, 2000–2002. [DOI] [PubMed] [Google Scholar]
- 8. Pennarun B, Meijer A, de Vries EGE, Kleibeuker JH, Kruyt F & de Jong S (2010) Playing the DISC: turning on TRAIL death receptor‐mediated apoptosis in cancer. Biochim Biophys Acta Rev Cancer 1805, 123–140. [DOI] [PubMed] [Google Scholar]
- 9. LeBlanc HN & Ashkenazi A (2003) Apo2L/TRAIL and its death and decoy receptors. Cell Death Differ 10, 66–75. [DOI] [PubMed] [Google Scholar]
- 10. Sheridan JP (1997) Control of TRAIL‐induced apoptosis by a family of signaling and decoy receptors. Science 277, 818–821. [DOI] [PubMed] [Google Scholar]
- 11. Kimberley FC & Screaton GR (2004) Following a TRAIL: update on a ligand and its five receptors. Cell Res 14, 359–372. [DOI] [PubMed] [Google Scholar]
- 12. Pan G (1997) An antagonist decoy receptor and a death domain‐containing receptor for TRAIL. Science 277, 815–818. [DOI] [PubMed] [Google Scholar]
- 13. Marsters SA, Sheridan JP, Pitti RM, Huang A, Skubatch M, Baldwin D, Yuan J, Gurney A, Goddard AD, Godowski P et al (1997) A novel receptor for Apo2L/TRAIL contains a truncated death domain. Curr Biol 7, 1003–1006. [DOI] [PubMed] [Google Scholar]
- 14. Clancy L, Mruk K, Archer K, Woelfel M, Mongkolsapaya J, Screaton G, Lenardo MJ & Chan FK‐M (2005) Preligand assembly domain‐mediated ligand‐independent association between TRAIL receptor 4 (TR4) and TR2 regulates TRAIL‐induced apoptosis. Proc Natl Acad Sci USA 102, 18099–18104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schneider P, Thome M, Burns K, Bodmer JL, Hofmann K, Kataoka T, Holler N & Tschopp J (1997) TRAIL receptors 1 (DR4) and 2 (DR5) signal FADD‐dependent apoptosis and activate NF‐κB. Immunity 7, 831–836. [DOI] [PubMed] [Google Scholar]
- 16. Chaudhary PM, Eby M, Jasmin A, Bookwalter A, Murray J & Hood L (1997) Death receptor 5, a new member of the TNFR family, and DR4 induce FADD‐dependent apoptosis and activate the NF‐κB pathway. Immunity 7, 821–830. [DOI] [PubMed] [Google Scholar]
- 17. Kuang AA, Diehl GE, Zhang J & Winoto A (2000) FADD is required for DR4‐ and DR5‐mediated apoptosis. Lack of trail‐induced apoptosis in FADD‐deficient mouse embryonic fibroblasts. J Biol Chem 275, 25065–25068. [DOI] [PubMed] [Google Scholar]
- 18. Siegmund D, Lang I & Wajant H (2017) Cell death‐independent activities of the death receptors CD95, TRAILR1, and TRAILR2. FEBS J 284, 1131–1159. [DOI] [PubMed] [Google Scholar]
- 19. Bodmer JL, Holler N, Reynard S, Vinciguerra P, Schneider P, Juo P, Blenis J & Tschopp J (2000) TRAIL receptor‐2 signals apoptosis through FADD and caspase‐8. Nat Cell Biol 2, 241–243. [DOI] [PubMed] [Google Scholar]
- 20. Sprick MR, Weigand MA, Rieser E, Rauch CT, Juo P, Blenis J, Krammer PH & Walczak H (2000) FADD/MORT1 and caspase‐8 are recruited to TRAIL receptors 1 and 2 and are essential for apoptosis mediated by TRAIL receptor 2. Immunity 12, 599–609. [DOI] [PubMed] [Google Scholar]
- 21. Kischkel FC, Lawrence DA, Chuntharapai A, Schow P, Kim KJ & Ashkenazi A (2000) Apo2L/TRAIL‐dependent recruitment of endogenous FADD and caspase‐8 to death receptors 4 and 5. Immunity 12, 611–620. [DOI] [PubMed] [Google Scholar]
- 22. Salvesen GS & Ashkenazi A (2011) SnapShot: Caspases. Cell 147, 476–476.e1. [DOI] [PubMed] [Google Scholar]
- 23. Budihardjo I, Oliver H, Lutter M, Luo X & Wang X (1999) Biochemical pathways of caspase activation during apoptosis. Annu Rev Cell Dev Biol 15, 269–290. [DOI] [PubMed] [Google Scholar]
- 24. Wagner KW, Punnoose EA, Januario T, Lawrence DA, Pitti RM, Lancaster K, Lee D, vonGoetz M , Yee SF, Totpal K et al (2007) Death‐receptor O‐glycosylation controls tumor‐cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med 13, 1070–1077. [DOI] [PubMed] [Google Scholar]
- 25. Saturno G, Valenti M, De Haven Brandon A, Thomas GV, Eccles S, Clarke PA & Workman P (2013) Combining trail with PI3 kinase or HSP90 inhibitors enhances apoptosis in colorectal cancer cells via suppression of survival signaling. Oncotarget 4, 1185–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kelley SK (2001) Preclinical studies to predict the disposition of Apo2L/tumor necrosis factor‐related apoptosis‐inducing ligand in humans: characterization of in vivo efficacy, pharmacokinetics, and safety. J Pharmacol Exp Ther 299, 31–38. [PubMed] [Google Scholar]
- 27. Trarbach T, Moehler M, Heinemann V, Köhne CH, Przyborek M, Schulz C, Sneller V, Gallant G & Kanzler S (2010) Phase II trial of mapatumumab, a fully human agonistic monoclonal antibody that targets and activates the tumour necrosis factor apoptosis‐inducing ligand receptor‐1 (TRAIL‐R1), in patients with refractory colorectal cancer. Br J Cancer 102, 506–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sirtl S, Knoll G, Trinh DT, Lang I, Siegmund D, Gross S, Schuler‐Thurner B, Neubert P, Jantsch J, Wajant H et al (2018) Hypertonicity‐enforced BCL‐2 addiction unleashes the cytotoxic potential of death receptors. Oncogene 37, 4122–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lim B, Scicchitano A, Beachler C, Gusani N, Sarwani N, Yang Z, Staveley‐O'Carroll K, Ashkenazi A, Portera C & El‐Deiry WS (2013) FOLFIRI plus dulanermin (rhApo2L/TRAIL) in a patient with BRAF‐mutant metastatic colon cancer. Cancer Biol Ther 14, 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tabernero J, Chawla SP, Kindler H, Reckamp K, Chiorean EG, Azad NS, Lockhart AC, Hsu CP, Baker NF, Galimi F et al (2015) Anticancer activity of the type I insulin‐like growth factor receptor antagonist, ganitumab, in combination with the death receptor 5 agonist, conatumumab. Target Oncol 10, 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Christiansen MN, Chik J, Lee L, Anugraham M, Abrahams JL & Packer NH (2014) Cell surface protein glycosylation in cancer. Proteomics 14, 525–546. [DOI] [PubMed] [Google Scholar]
- 32. Becker DJ & Lowe JB (2003) Fucose: Biosynthesis and biological function in mammals. Glycobiology 13, 41R–53R. [DOI] [PubMed] [Google Scholar]
- 33. Miyoshi E, Moriwaki K & Nakagawa T (2008) Biological function of fucosylation in cancer biology. J Biochem 143, 725–729. [DOI] [PubMed] [Google Scholar]
- 34. Haltiwanger RS (2009) Fucose is on the TRAIL of colon cancer. Gastroenterology 137, 36–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moriwaki K, Noda K, Furukawa Y, Ohshima K, Uchiyama A, Nakagawa T, Taniguchi N, Daigo Y, Nakamura Y, Hayashi N et al (2009) Deficiency of GMDS leads to escape from NK cell‐mediated tumor surveillance through modulation of TRAIL signaling. Gastroenterology 137, 188–198. [DOI] [PubMed] [Google Scholar]
- 36. Moriwaki K, Narisada M, Imai T, Shinzaki S & Miyoshi E (2010) The effect of epigenetic regulation of fucosylation on TRAIL‐induced apoptosis. Glycoconj J 27, 649–659. [DOI] [PubMed] [Google Scholar]
- 37. Dufour F, Rattier T, Shirley S, Picarda G, Constantinescu AA, Morlé A, Zakaria AB, Marcion G, Causse S, Szegezdi E et al (2017) N‐glycosylation of mouse TRAIL‐R and human TRAIL‐R1 enhances TRAIL‐induced death. Cell Death Differ 24, 500–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van der Sloot AM, Tur V, Szegezdi E, Mullally MM, Cool RH, Samali A, Serrano L & Quax WJ (2006) Designed tumor necrosis factor‐related apoptosis‐inducing ligand variants initiating apoptosis exclusively via the DR5 receptor. Proc Natl Acad Sci USA 103, 8634–8639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reis CR, van der Sloot AM, Natoni A, Szegezdi E, Setroikromo R, Meijer M, Sjollema K, Stricher F, Cool RH, Samali A et al (2010) Rapid and efficient cancer cell killing mediated by high‐affinity death receptor homotrimerizing TRAIL variants. Cell Death Dis 1, e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Duiker EW, De Vries EGE, Mahalingam D, Meersma GJ, Van Ek WB, Hollema H, De Hooge MNL, Van Dam GM, Cool RH, Quax WJ et al (2009) Enhanced antitumor efficacy of a DR5‐specific TRAIL variant over recombinant human TRAIL in a bioluminescent ovarian cancer xenograft model. Clin Cancer Res 15, 2048–2057. [DOI] [PubMed] [Google Scholar]
- 41. Meijer A, Kruyt FAE, van der Zee AGJ, Hollema H, Le P, ten Hoor KA, Groothuis GMM, Quax WJ, de Vries EGE & de Jong S (2013) Nutlin‐3 preferentially sensitises wild‐type p53‐expressing cancer cells to DR5‐selective TRAIL over rhTRAIL. Br J Cancer 109, 2685–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Szegezdi E, van der Sloot AM, Mahalingam D, O'Leary L, Cool RH, Munoz IG, Montoya G, Quax WJ, de Jong S, Samali A et al (2012) Kinetics in signal transduction pathways involving promiscuous oligomerizing receptors can be determined by receptor specificity: apoptosis induction by TRAIL. Mol Cell Proteomics 11, M111.013730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moriwaki K, Shinzaki S & Miyoshi E (2011) GDP‐mannose‐4,6‐dehydratase (GMDS) deficiency renders colon cancer cells resistant to tumor necrosis factorrelated apoptosis‐inducing ligand (TRAIL) receptor‐ and CD95‐mediated apoptosis by inhibiting complex II formation. J Biol Chem 286, 43123–43133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Burkart MD, Vincent SP, Düffels A, Murray BW, Ley SV & Wong CH (2000) Chemo‐enzymatic synthesis of fluorinated sugar nucleotide: useful mechanistic probes for glycosyltransferases. Bioorganic Med Chem 8, 1937–1946. [DOI] [PubMed] [Google Scholar]
- 45. Rillahan CD, Antonopoulos A, Lefort CT, Sonon R, Ley K, Dell A, Haslam SM & Paulson JC (2012) Global metabolic inhibitors of sialyl‐ and fucosyltransferases. Nat Chem Biol 8, 661–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fukumori F, Takeuchi N, Hagiwara T, Ohbayashi H, Endo T, Kochibe N, Nagata Y & Kobata A (1990) Primary structure of a fucose‐specific lectin obtained from a mushroom, Aleuria aurantia . J Biochem 107, 190–196. [DOI] [PubMed] [Google Scholar]
- 47. Wimmerova M, Mitchell E, Sanchez J, Gautier C & Imberty A (2003) Crystal structure of fungal lectin six‐bladed beta‐propeller fold and novel fucose recognition mode for Aleuria aurantia lectin. J Biol Chem 278, 27059–27067. [DOI] [PubMed] [Google Scholar]
- 48. Sessler T, Healy S, Samali A & Szegezdi E (2013) Structural determinants of DISC function: new insights into death receptor‐mediated apoptosis signalling. Pharmacol Ther 140, 186–199. [DOI] [PubMed] [Google Scholar]
- 49. Polanski R, Vincent J, Polanska UM, Petreus T & Tang EKY (2015) Caspase‐8 activation by TRAIL monotherapy predicts responses to IAPi and TRAIL combination treatment in breast cancer cell lines. Cell Death Dis 6, e1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu Y‐C, Yen H‐Y, Chen C‐Y, Chen C‐H, Cheng P‐F, Juan Y‐H, Chen C‐H, Khoo K‐H, Yu C‐J, Yang P‐C et al (2011) Sialylation and fucosylation of epidermal growth factor receptor suppress its dimerization and activation in lung cancer cells. Proc Natl Acad Sci USA 108, 11332–11337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Marquardt T, Lühn K, Srikrishna G, Freeze HH, Harms E & Vestweber D (1999) Correction of leukocyte adhesion deficiency type II with oral fucose. Blood 94, 3976–3985. [PubMed] [Google Scholar]
- 52. Lühn K, Marquardt T, Harms E & Vestweber D (2001) Discontinuation of fucose therapy in LADII causes rapid loss of selectin ligands and rise of leukocyte counts. Blood 97, 330–332. [DOI] [PubMed] [Google Scholar]
- 53. Tomsik P, Soukup T, Cermakova E, Micuda S, Niang M, Sucha L & Rezacova M (2011) L‐rhamnose and L‐fucose suppress cancer growth in mice. Cent Eur J Biol 6, 1–9. [Google Scholar]
- 54. Neumann S, Hasenauer J, Pollak N & Scheurich P (2014) Dominant negative effects of tumor necrosis factor (tnf)‐related apoptosis‐inducing ligand (TRAIL) receptor 4 on TRAIL receptor 1 signaling by formation of heteromeric complexes. J Biol Chem 289, 16576–16587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Aoyagi Y, Suzuki Y, Igarashi K, Saitoh A, Oguro M, Yokota T, Mori S, Nomoto M, Isemura M & Asakura H (1991) The usefulness of simultaneous determinations of glucosaminylation and fucosylation indices of alpha‐fetoprotein in the differential diagnosis of neoplastic diseases of the liver. Cancer 67, 2390–2394. [DOI] [PubMed] [Google Scholar]
- 56. Taketa K, Endo Y, Sekiya C, Tanikawa K, Koji T, Taga H, Satomura S, Matsuura S, Kawai T & Hirai H (1993) A collaborative study for the evaluation of lectin‐reactive a‐fetoproteins in early detection of hepatocellular carcinoma. Cancer Res 53, 5419–5424. [PubMed] [Google Scholar]
- 57. Kyselova Z, Mechref Y, Kang P, Goetz JA, Dobrolecki LE, Sledge GW, Schnaper L, Hickey RJ, Malkas LH & Novotny MV (2008) Breast cancer diagnosis and prognosis through quantitative measurements of serum glycan profiles. Clin Chem 54, 1166–1175. [DOI] [PubMed] [Google Scholar]
- 58. Lin Z, Simeone D, Anderson MA, Brand R, Xie X, Shedden K, Ruffin MT, Lubman DM, Shedden KA & Lubman DM (2011) Mass spectrometric assay for analysis of haptoglobin fucosylation in pancreatic cancer. J Proteome Res 10, 2602–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]


