Abstract
The Notch pathway plays diverse and complex roles in cell signaling during development. In the mammalian ovary, Notch is important for the initial formation and growth of follicles, and for regulating the proliferation and differentiation of follicular granulosa cells during the periovulatory period. This study seeks to determine the contribution of female germ cells toward the initial activation and subsequent maintenance of Notch signaling within somatic granulosa cells of the ovary. To address this issue, transgenic Notch reporter (TNR) mice were crossed with Sohlh1-mCherry (S1CF) transgenic mice to visualize Notch-active cells (EGFP) and germ cells (mCherry) simultaneously in the neonatal ovary. To test the involvement of oocytes in activation of Notch signaling in ovarian somatic cells, we ablated germ cells using busulfan, a chemotherapeutic alkylating agent, or investigated KitWv/Wv (viable dominant white-spotting) mice that lack most germ cells. The data reveal that Notch pathway activation in granulosa cells is significantly suppressed when germ cells are reduced. We further demonstrate that disruption of the gene for the Notch ligand Jag1 in oocytes similarly impacts Notch activation and that recombinant JAG1 enhances Notch target gene expression in granulosa cells. These data are consistent with the hypothesis that germ cells provide a ligand, such as Jag1, that is necessary for activation of Notch signaling in the developing ovary.
The ovary is important for female fertility, as it contributes to reproductive health through the production of sex hormones and the generation of follicles that facilitate oocyte development (1, 2). The earliest follicles are composed of two cell types, the oocyte and the pregranulosa cells, that interact during a process termed nest breakdown (1, 2) in which germ cells connected by cytoplasmic bridges are invaded by pregranulosa cells to encapsulate individual oocytes. Selected cohorts of newly formed primordial follicles are then recruited to undergo growth and maturation following sexual maturity (3). The establishment of a finite number of primordial follicles during the perinatal period is important, as these follicles represent the reproductive potential of the female organism. Although there are multiple signaling modalities that are necessary for the development of the follicle (4), there has been recent focus on juxtacrine, or contact-dependent, signaling because of the spatial relationships and interactions between the oocyte and the surrounding somatic pregranulosa cells (5, 6).
Studies investigating juxtacrine signaling, specifically Notch signaling, have shown that this pathway is involved in follicle development and overall female fertility (7–15). There is an activation of Notch signaling in the ovary during the time of germ cell nest breakdown and follicle establishment starting at embryonic day (E)15.5 in the mouse (8). Notch activity, as measured using the transgenic Notch reporter (TNR) (16), an EGFP reporter gene expressed dependent on the Notch pathway transcription factor Rbpj, increases throughout embryonic development and continues postnatally during follicle growth (8). Notch activity is observed at postnatal day (PND)0 in somatic cells, identified as granulosa cells, that form intricate cage-like structures that encircle oocytes (8). Quantitative gene expression analyses using whole ovaries revealed significant expression of Notch component and downstream effector mRNAs, with Notch2, Jagged1, and Hes1 being particularly abundantly expressed at embryonic (8) and postnatal times (10). Additionally, in situ hybridization and immunolocalization studies showed that the receptors Notch2 (8, 10) and Notch3 (9, 17) are expressed in granulosa cells, the ligand Jagged1 (8, 10) is expressed in oocytes, and the ligand Jagged2 (9, 18) is expressed in both cell types depending on the follicle stage studied. With this dynamic temporal expression of Notch components, as well as the observed spatial relationships between Notch receptors and ligands, Notch signaling has the potential to play roles in cell-to-cell communication and the regulation of follicle and ovarian function.
Experiments to inhibit Notch signaling in the ovary have revealed several reproductive phenotypes. Following Notch inhibition with the γ-secretase inhibitor N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT) (19), cultured ovaries showed increased retention of germ cell nests and decreased primordial follicle populations (10, 20, 21). Additionally, ovaries in which Notch activity was inhibited were shown to have more apoptotic cells, suggesting that Notch contributes to granulosa cell survival in newly formed follicles (10, 20, 21). Mice with conditional Jagged1 knockout (cJ1KO) or conditional Notch2 knockout (cN2KO) within the oocytes or granulosa cells, respectively, have reproductive phenotypes impacting follicle formation and growth. These include a decrease in the primordial follicle population and an increased incidence of multi-oocytic and abnormal follicles, although many normal follicles remain (8, 15). Granulosa cells within follicles in these mice also have increased apoptosis and decreased proliferation, although not all follicles are impacted and some mature normally (8). Furthermore, gene expression analyses reveal decreased expression of Notch components and downstream effectors (8). The phenotypes observed within these conditional knockout mice were correlated with altered fertility. Our group showed that cJ1KO mice were subfertile (8), whereas Xu and Gridley (15) found that the cN2KO mice generated in their laboratory were likewise subfertile. Overall, these data highlight the importance of Notch signaling in follicle development and ovarian function.
Based on the localization of Notch ligands in the oocyte and Notch receptors in the surrounding granulosa cells, we hypothesized that the oocyte may be an important source of ligand for initial activation of Notch signaling in the pregranulosa cells and that reducing the oocyte population during development would affect Notch activation in ovarian pregranulosa cells. To test this, we used two mouse models with a reduced oocyte pool and investigated the consequences on Notch signaling either during late embryogenesis or postnatally. We first ablated oocytes using a chemotherapeutic agent, busulfan (22, 23), in a double-reporter mouse that fluorescently labeled Notch-active cells using the TNR (16) and germ cells using a Sohlh1-mCherry (S1CF) reporter (24). Because we found that busulfan-treated mice were unable to deliver viable pups for postnatal analysis, we also used a mouse with a point mutation of the c-Kit receptor, an important component of KIT signaling that impacts oocyte migration, proliferation, and survival (25, 26). The KitWv/Wv or viable dominant white-spotting mouse (25), expressing the same Notch activity reporter, was used to study differences in Notch activation and distribution in early postnatal development. We demonstrated that a reduction of oocytes is associated with decreased Notch activity, although a basal level of reporter expression remains. To test whether the Notch ligand JAG1 from oocytes might act on Notch receptors in granulosa cells, we examined oocyte-specific Jag1 knockout mice and found decreased expression of the Notch transgenic reporter. Finally, we complement our in vivo studies by demonstrating that recombinant JAG1 activates Notch target genes in cultured granulosa cells, as well as inducing its own expression, thus providing a potential mechanism of signal propagation in growing follicles. These results are consistent with the oocyte and JAG1 being important for initial Notch pathway activation in the developing mouse ovary.
Methods
Mouse care and generation of mouse lines
Mice were housed in controlled environmental conditions with access to water and food ad libitum on a 12-hour light/12-hour dark cycle. Mice were fed a diet free of alfalfa and soybean meal to minimize levels of naturally occurring phytoestrogens and to reduce autofluorescence in tissue samples used for ex vivo imaging (2919 Teklad diet for breeding and 2916 Teklad diet for maintenance, Harlan Laboratories, Indianapolis, IN). Timed matings were used, with E0.5 designated as 12:00 pm on the day of vaginal plug detection. PND0 was designated as the first 24 hours after birth. All procedures were approved by the Northwestern University Institutional Animal Care and Use Committee.
The TNR and SOHLH1-mCherry (S1CF) reporter lines (27) were graciously provided by Dr. Nicholas Gaiano from Johns Hopkins University (16) and Dr. Aleksandar Rajkovic from University of Pittsburgh (24), respectively. Female TNR/+ mice were crossed with male S1CF/+ mice to generate TNR/+; S1CF/+ mice. Male TNR/+; S1CF/+ mice were crossed with wild-type (WT) females to generate TNR/+; S1CF/+ embryos. Genotyping was performed by PCR using primer sets for EGFP and mCherry; primer sequences can be found in an online repository (27).
The KitWv/Wv mouse (25) carrying the TNR Notch reporter was generated through a cross between a KitWv/+ mouse, provided by the laboratory of Danielle Maatouk at Northwestern University (28), and a TNR/+ mouse. After confirmation of the TNR/+; Kit Wv/+ heterozygous mouse genotype, these mice were crossed with heterozygous KitWv/+ mice to generate homozygous TNR/+; KitWv/Wv mice. Primer sequences for genotyping can be found in an online repository (27).
The cJ1KO mouse model was generated as previously described (8). The TNR reporter was incorporated into the cJ1KO mouse line for these studies.
Busulfan treatment
TNR/+; S1CF/+ male mice were mated with CD-1 females until a vaginal plug was detected. The pregnant dams were monitored until 11.5 days after conception, when the mice were injected IP with vehicle or busulfan (100 mg/kg) dissolved in 90% corn oil/10% ethanol. The pregnant dam was monitored until 18.5 days after conception (E18.5), when intact embryos were extracted for embryonic ovary isolation. Isolated ovaries were processed for histology, imaging, and gene expression analyses.
Primary granulosa cell culture and recombinant JAG1 ligand
To make recombinant JAG1–coated substrate, tissue culture plates were incubated with a solution of 5 µg/mL recombinant rat Jagged1-Fc chimeric protein (R&D Systems, Minneapolis, MN) and 1 µg/cm2 fibronectin (Sigma-Aldrich, St. Louis, MO) overnight at 4°C with shaking to allow for protein adsorption. Control wells were coated with fibronectin only. Ovaries were dissected from PND19 mice. Granulosa cells were collected by follicle puncture and cultured as previously described (29). Oocytes were removed with a 40-μm cell strainer (Fisher Scientific, Hampton, NH). Granulosa cells were plated onto the control or recombinant JAG1 substrates at 175,000 cells per well in 24-well plates in a humidified incubator at 37°C and 5% CO2 using a 1:1 ratio of DMEM/F12 medium (Fisher Scientific) supplemented with 15 mM HEPES (pH 7.4), 5 mg/mL transferrin, 2 mg/mL insulin, 40 ng/mL hydrocortisone, 10% fetal bovine serum, and 100 U/mL penicillin/streptomycin (4F media). Cells were allowed to adhere overnight, followed by treatment with 20 μM DAPT (SelleckChem, Houston, TX) or DMSO vehicle for 24 hours in those experiments where an inhibitor was used.
Quantitative reverse transcription PCR gene expression analysis
Ovaries were isolated and preserved in RNAlater reagent (Life Technologies, Carlsbad, CA) at −80°C until RNA extraction. RNA was extracted using an RNeasy Plus mini kit (Zymo Research, Irvine, CA). RNA concentration and quality were assessed using a NanoDrop spectrophotometer (Thermo Scientific, Carlsbad, CA). RNA was reverse transcribed to cDNA using SuperScript VILO master mix (Life Technologies). Quantitative reverse transcription PCR (qRT-PCR) assays were performed using SYBR Green PCR master mix (Life Technologies) with an Applied Biosystems 7300 (Life Technologies) thermocycler. The comparative cycle threshold method (30) was implemented for relative quantification using Rpl19 as an internal control (31). The sequences for primers used in gene expression analyses can be found in an online repository (27).
Confocal microscopy
Ovaries were dissected at the specified time points and placed in PBS, stained with Hoechst 33234 dye (10 mg/mL) for 15 minutes, and imaged to detect the EGFP and mCherry fluorescent reporters. Confocal imaging of whole ovaries was performed using either a Leica SP5 confocal microscope or a Leica SP8 confocal microscope with the following filters: 401 nm for Hoechst 33234, 488 nm for EGFP, and 536 nm for mCherry. Images were processed from Z-stack data (1-μm step size) of the entire gonad at PND0 and PND3, and of the middle 70 μm of the gonad at P10 to generate a maximum intensity projection of the fluorescent channels using Fiji/ImageJ (32).
Quantification of EGFP fluorescence
Five independent 50- × 50-μm squares were randomly placed on confocal images from control or busulfan-treated ovaries using an ImageJ grid generator. The mean fluorescence intensity of each EGFP-positive cell within the squares was measured, also using ImageJ. For experiments to establish proximity relationships between oocytes and EGFP-positive cells, mCherry-positive oocytes were identified (also within randomly placed squares, as above), and EGFP-positive cells within that square were scored as being either in contact with a circle of diameter 25 μm surrounding that oocyte (∼20 μm), or outside of this circle. Finally, for the busulfan-treated group, the mean fluorescence intensity was determined for individual EGFP-positive cells either adjacent to oocytes or not, using the radial distance method described above. Four independent mice from each treatment group were analyzed.
Histological examination and immunohistochemistry
Ovarian tissue samples were dissected and fixed overnight at 4°C in 4% paraformaldehyde in PBS and dehydrated in 70% ethanol for storage. Samples were embedded in paraffin and sectioned at 5 µm for histological analysis. Hematoxylin and eosin staining, immunohistochemistry, and immunofluorescence were performed as described in previous publications (8, 10).
Statistical analysis
Data are presented as means ± SEM. Experiments were performed using replicates and control groups as stated in the figure legends. An F test was conducted to compare homogeneity of variances between the relevant treatment groups or genotypes. When an F test indicated a significant difference in variances between the means, nonparametric testing was conducted as appropriate. Differences between groups were calculated using GraphPad Prism (GraphPad Software, La Jolla, CA) applying a two-tailed t test with Bonferroni correction, or ANOVA with a Tukey post hoc test, as appropriate. Differences are indicated as significant at a 95% CI (P < 0.05).
Results
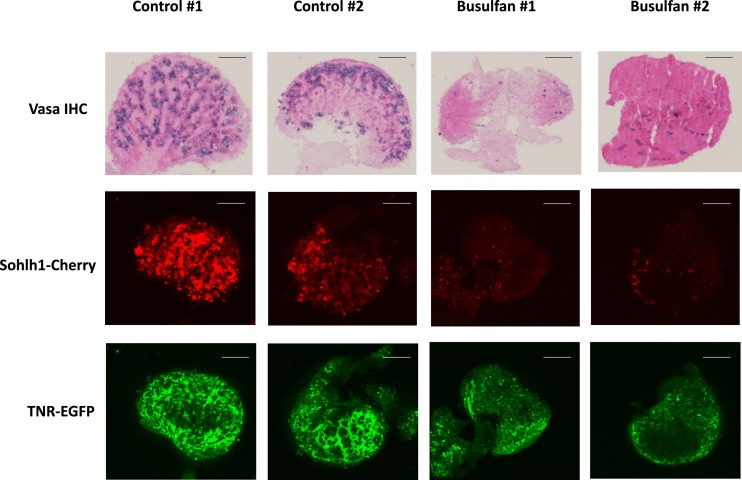
Busulfan-treated and control TNR/+; S1CF/+ E18.5 ovaries were assessed qualitatively for oocytes and for EGFP expression using immunodetection as well as confocal imaging. E18.5 was chosen to allow significant time for busulfan, administered at E11.5, to exert its effects while recognizing that the busulfan-treated dams are unable to give birth. Vasa immunodetection (33) and mCherry fluorescence were both decreased in the busulfan-injected ovaries, confirming a reduction in the number of oocytes (Fig. 1). Imaging of the busulfan-treated ovaries also revealed an overall decrease in EGFP fluorescence compared with controls. Both the mCherry and EGFP signals were still detected in the busulfan-treated ovaries, suggesting that some germ cells remained, as well as some residual Notch activity (Fig. 1).
Figure 1.
Imaging TNR (EGFP) expression following busulfan treatment of embryonic TNR/S1CF ovaries. E18.5 ovaries isolated from dams injected with vehicle (control) or busulfan at gestation day E11.5 are shown. Whole ovaries were used for confocal fluorescence microscopy to detect the Sohlh1-mCherry reporter or the TNR-EGFP reporter and were subsequently sectioned and processed for immunohistochemical detection of VASA protein. Original magnification, ×20; scale bars, 50 µm. Images from two representative animals are shown from n = 5 for each condition.
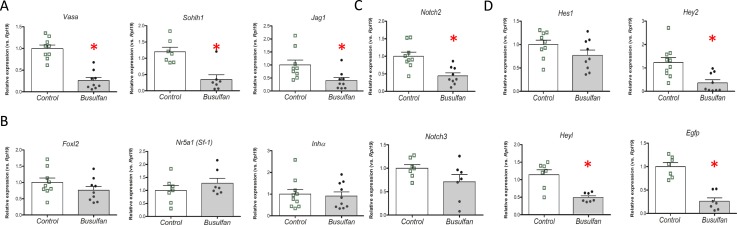
We quantitatively evaluated the effects of busulfan by examining mRNA expression of oocyte- and granulosa cell–specific genes. Consistent with the imaging, oocyte-specific Vasa, Sohlh1, and Jag1 mRNA expression was found to be significantly decreased (Fig. 2A), whereas the granulosa cell markers Nr5a1 (SF-1), Foxl2, and Inhα showed no significant changes in mRNA abundance (Fig. 2B). Expression of the mRNAs for the most abundant Notch receptors in granulosa cells, Notch2 and Notch3 (8, 10), was also examined. Notch2 was significantly reduced, whereas Notch3 was unchanged (Fig. 2C). These data confirm that busulfan depleted a significant fraction of the oocyte population, but the expression of multiple genes specific to granulosa cells within the ovary was not significantly affected by the drug.
Figure 2.
Ovarian gene expression changes following embryonic busulfan exposure. Ovaries were isolated at E18.5 from TNR/+; S1CF/+ embryos from dams injected at E11.5 with vehicle or busulfan, and RNA was prepared for qRT-PCR analyses of mRNA abundance. (A) Germ cell markers. (B) Granulosa cell markers. (C) Notch receptors. (D) Notch target/effector and reporter genes. n = 7 to 10 animals, with each animal represented as either an open square (control) or filled circle (busulfan) in the scatter plots. Means ± SEM are shown. *P < 0.05.
Notch downstream target/effector gene expression and Notch reporter activity were next analyzed by measuring Hes1, Hey2, Heyl, and Egfp mRNAs. Both Hey2 and Heyl mRNAs were decreased in the busulfan-treated samples, although Hes1 was unchanged (Fig. 2D). Most importantly, the Notch activity reporter, the TNR Egfp mRNA, showed a significant decrease in expression in busulfan-treated ovaries (Fig. 2D). This suggests that the substantial reduction in oocytes caused by busulfan treatment negatively affects overall Notch activity in the embryonic ovary.
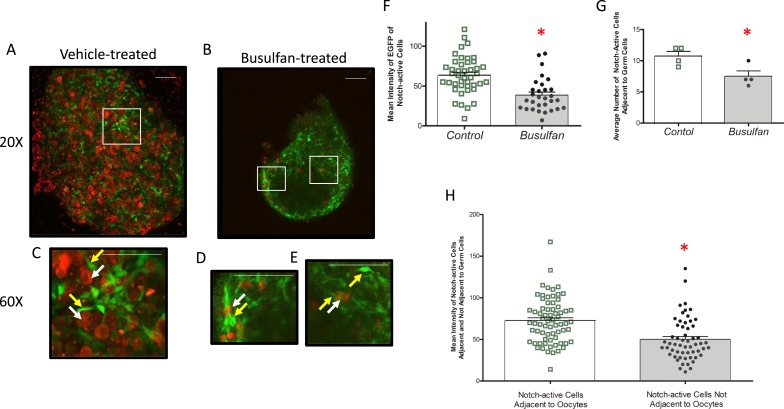
Due to the residual Notch activity observed in the busulfan-treated ovary, we sought to investigate the spatial relationships between remaining oocytes and Notch-active cells. At E18.5, oocytes that remained following busulfan treatment and expressed the S1CF transgene were surrounded by strongly positive EGFP-expressing somatic cells (Fig. 3). Areas devoid of oocytes also had EGFP-expressing cells, but they were in general not as abundant or intense. Similarly, there were rare oocytes not associated with EGFP-expressing cells, although the health of these oocytes following the busulfan treatment is not known. To more quantitatively evaluate localization and expression of the TNR reporter in these ovaries, the mean fluorescence intensities and numbers of EGFP-expressing cells in both the control and busulfan-treated groups were determined, as well as the mean fluorescence intensities of somatic cells either adjacent to, or away from, oocytes following busulfan treatment (Fig. 3). These data support a relationship between oocytes and those somatic cells that most intensely express the EGFP Notch reporter.
Figure 3.
Relationship between Notch activity and remaining germ cells in the busulfan-exposed embryonic ovary. Embryonic ovaries isolated at E18.5 from dams injected with (A) vehicle or (B) busulfan at E11.5 are shown. Whole ovaries were used for confocal fluorescence microscopy to detect the Sohlh1-mCherry reporter (germ cells) or the TNR-EGFP reporter (Notch-active cells). The white boxes indicate specific areas shown at increased magnification in (C)–(E). The white arrows indicate oocytes, and the yellow areas show GFP-positive somatic cells. These are generally in close association (C and D), although examples of GFP-positive cells without an apparent oocyte in the visual field are observed [single yellow arrow in (E)]. (F) Mean fluorescence intensity of EGFP-expressing somatic cells in control and busulfan-treated ovaries is shown, determined as described in the “Methods” section. (G) The average number of these EGFP-positive cells in proximity to oocytes as a function of the treatment is shown. (H) Mean fluorescence intensity of EGFP-positive cells either adjacent to or away from oocytes, determined as described in the “Methods” section. n = 4 mice for each group. (A–E) Scale bars, 50 μm. The bars over the scatter plots designate mean ± SEM. *P < 0.05.
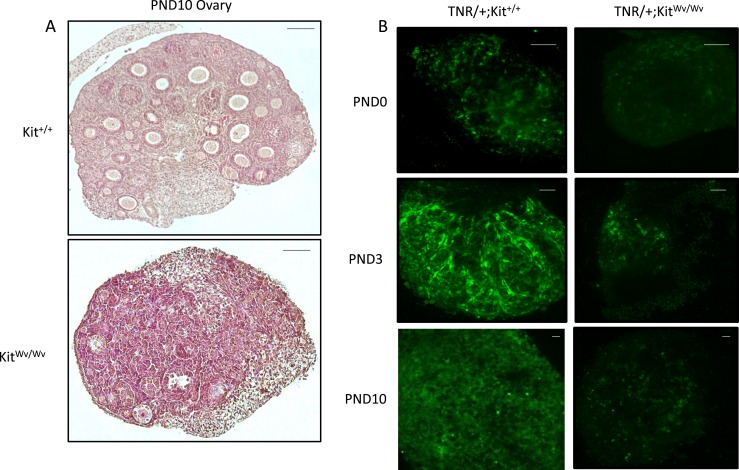
To examine postnatal ovaries, we used a Kit mutant mouse line (KitWv/Wv) (25), known as the dominant white-spotting mouse, in which a point mutation in the c-Kit receptor suppresses oocyte migration, proliferation, and viability (26). Ovaries from KitWv/Wv mice are smaller in size and have a very limited oocyte population compared with controls, making them useful in studying the relationship between oocytes and Notch activity. We did not use the Sohlh1-mCherry reporter at these postnatal times because it does not mark oocytes beyond the primordial follicle stage (24, 34). The Notch TNR reporter was introduced into KitWv/Wv mice, and oocyte depletion was confirmed by histologically examining PND10 ovaries (Fig. 4A). TNR/+; Kit+/+ and TNR/+; KitWv/Wv ovaries were imaged to assess EGFP expression and distribution at PND0, PND3, and PND10, times associated with the germ cell nest stage (PND0), germ cell nest breakdown to form primordial follicles (PND3), and initial growth of primary and early secondary follicles (PND10) (Fig. 4B). At each time point, EGFP fluorescence signal in the KitWv/Wv ovaries was decreased compared with controls.
Figure 4.
TNR (EGFP) expression in the tnr/white-spotted mouse ovary. (A) Ovarian histology of representative control (Kit+/+) and mutant (KitWv/Wv) animals at PND10. Scale bars, 100 µm. (B) EGFP imaging at PND0, PND3, and PND10 in control and white-spotted ovaries. Original magnification, ×20; scale bars, 100 µm. These are representative of n = 3 for each group at each time point shown.
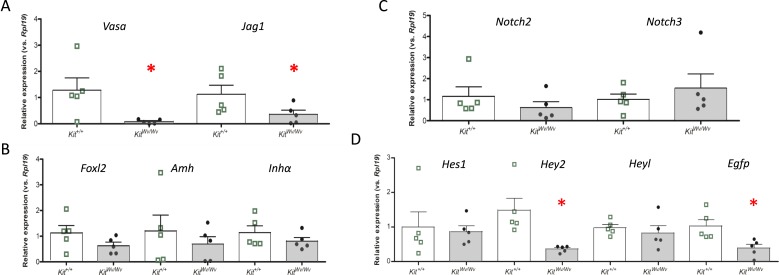
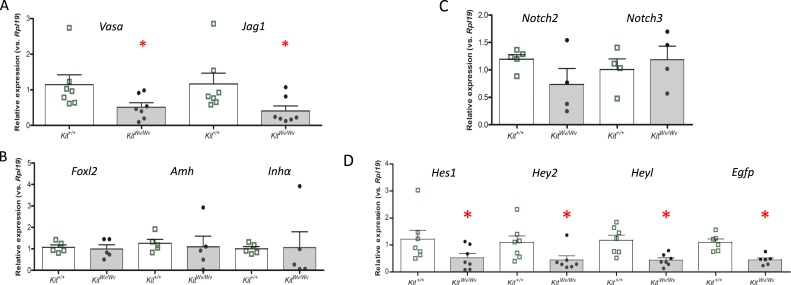
Quantitative gene expression analyses were conducted as described for the busulfan studies using the oocyte-specific factors Vasa and Jag1, as well as the granulosa cell-specific factors, Foxl2, Amh, and Inhα. Fig. 5 shows data from PND0, and Fig. 6 shows data from PND19. Vasa and Jag1 mRNA expression were significantly decreased in KitWv/Wv mice in comparison with Kit+/+ mice (Figs. 5A and 6A), although the three granulosa cell markers showed no significant changes (Figs. 5B and 6B). The Notch receptors Notch2 and Notch3 were likewise unchanged (Figs. 5C and 6C). Next, Notch target/effector gene expression was investigated. At PND0 Hey2 was decreased (Fig. 5D), and by PND19 Hey1, Hey2, and Heyl were all decreased (Fig. 6D). Finally, the EGFP Notch activity reporter was significantly repressed at both postnatal times (Figs. 5D and 6D). Similar results were obtained at PND3 and PND10 (27). Altogether, imaging and gene expression analyses of the oocyte-depleted KitWv/Wv ovary revealed significantly attenuated Notch activity throughout prepubertal postnatal development.
Figure 5.
Ovarian gene expression in control and white-spotted mouse ovaries at PND0. Ovaries were isolated at PND0 from TNR/Kit+/+ mice (open bars with green boxes) or TNR/KitWv/Wv mice (filled bars with black circles) and RNA was prepared for qRT-PCR analyses of mRNA abundance. (A) Germ cell markers. (B) Granulosa cell markers. (C) Notch receptors. (D) Notch target/effector and reporter genes. n = 5 animals, with each animal represented as a point in the scatter plots. The bars designate mean ± SEM. *P < 0.05.
Figure 6.
Ovarian gene expression in control and white-spotted mouse ovaries at PND19. Ovaries were isolated at PND19 from TNR/Kit+/+ mice (open bars with green boxes) or TNR/KitWv/Wv mice (filled bars with black circles) and RNA was prepared for qRT-PCR analyses of mRNA abundance. (A) Germ cell markers. (B) Granulosa cell markers. (C) Notch receptors. (D) Notch target/effector and reporter genes. n = 4 to 7, with the number of animals for each measurement indicated by the individual symbols shown in the scatter plots. The bars designate the mean ± SEM. *P < 0.05.
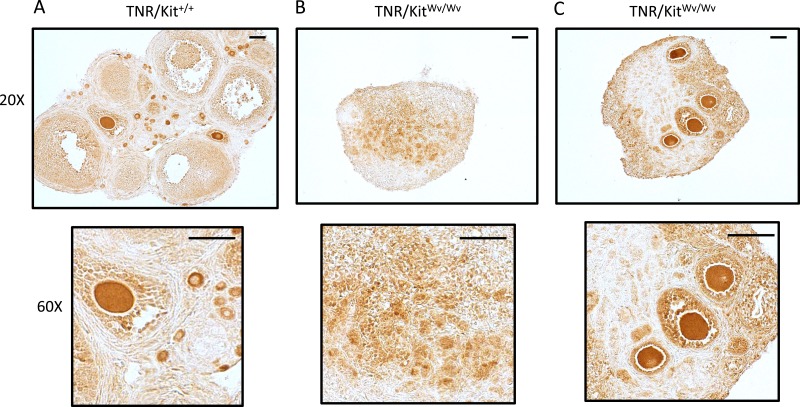
Whereas there are almost no oocytes in the KitWv/Wv ovaries, a small number do survive in some animals (26). To investigate spatial relationships between Notch-active cells and any surviving oocytes, immunodetection using an EGFP antibody (35) was performed on TNR/Kit+/+ and TNR/KitWv/Wv ovaries at PND19 (Fig. 7). There was strong EGFP immunostaining within control samples, particularly within granulosa cells adjacent to oocytes (Fig. 7A). In contrast, EGFP expression in KitWv/Wv ovaries varied, depending on the presence of oocytes. In the absence of oocytes (Fig. 7B) there was a detectable but diffuse signal observed. However, in the presence of oocytes that had survived (Fig. 7C) there was an intense and localized EGFP signal observed in the granulosa cells of follicles that had formed around these oocytes. The oocytes that remained in the PND19 Kit-mutant ovaries are not fragmented, appear morphologically healthy, and have a size and shape appropriate to the follicle. However, others have demonstrated that by 6 to 8 weeks of age these oocytes are lost as the ovary becomes devoid of all germ cells (26).
Figure 7.
TNR (EGFP) expression and localization in control and white-spotted mouse ovaries at PND19. (A) TNR/Kit+/+ ovary and (B) TNR/KitWv/Wv ovaries. PND19 ovaries were used for immunohistochemical detection of the EGFP Notch reporter. Original magnifications, ×20 and ×60; scale bars, 50 µm. These are representative images from n = 3 mice. The two TNR/KitWv/Wv ovaries shown illustrate one example without any apparent oocytes (left) and another with multiple remaining oocytes (right).
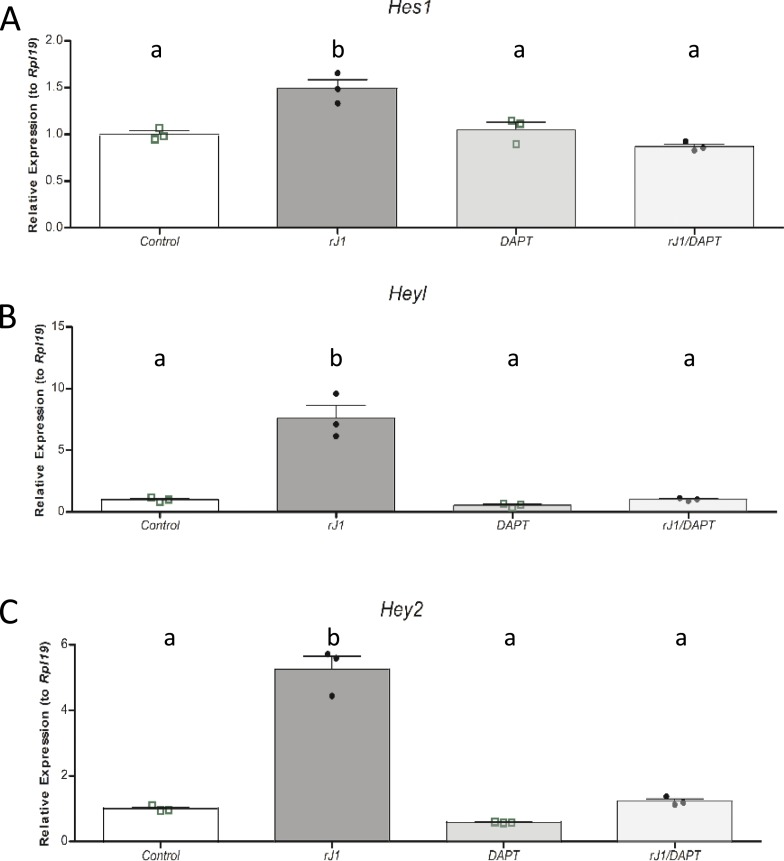
The observations in both the busulfan-treated and KitWv/Wv mouse models support the concept that the oocyte serves as an important signal for Notch pathway activation in the surrounding somatic cells during ovarian development. Prime candidates for this signal are the Notch ligands Jag1 and Jag2. We elected to focus on Jag1, as it is more abundantly expressed in the ovary and is largely restricted to the oocyte at these prepubertal times (8, 9). To ask whether Jag1 is sufficient to activate the Notch pathway in granulosa cells, we turned to cultured primary granulosa cells where it was possible to test the effects of an rJAG1 fusion protein immobilized to the culture dish (36–38). Indeed, we observed that recombinant JAG1 stimulated expression of the Notch target/effector genes Hes1, Hey2, and Heyl (Fig. 8). This effect was specific, in that it was reversed in the presence of the pan-Notch inhibitor DAPT.
Figure 8.
Effects of recombinant Jag1 protein on Notch target gene expression in granulosa cells. Primary cultured granulosa cells were plated onto immobilized recombinant JAG1 (rJ1) overnight as described in the “Methods” section and treated with vehicle or the Notch inhibitor DAPT for 24 h before isolation of RNA. Expression of the Notch target/effector genes (A) Hes1, (B) Hey2, and (C) Heyl was analyzed. n = 3 for each condition, with each biological replicate indicated as a point on the scatter plots. The bars designate mean ± SEM. Means not sharing the same letter are significantly different (at P < 0.05).
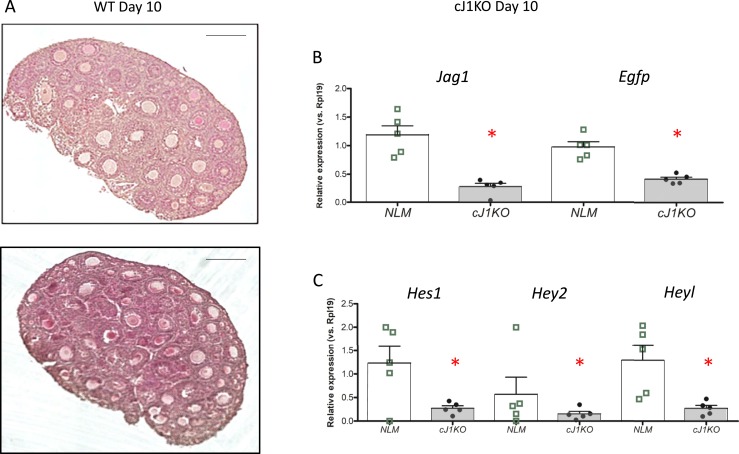
To ask whether Jag1 is necessary for the observed effects of oocytes on Notch activation, we turned to mice in which the Jag1 gene is conditionally disrupted in oocytes (8). Although there are follicular abnormalities in these ovaries, particularly apparent at later stages of development (8), at PND10 they have abundant oocytes within normal-appearing follicles (Fig. 9A). To investigate Notch activation, we crossed mice carrying the TNR Notch reporter into the cJ1KO background (TNR/Jag1−/− mice). Ovaries were harvested at PND10 for mRNA expression analysis. Jag1 mRNA is significantly reduced as expected, as is the Egfp Notch reporter mRNA (Fig. 9B) and the endogenous Notch target gene mRNAs Hes1, Hey2, and Heyl (Fig. 9C). Thus, in the presence of oocytes, loss of Jag1 from the oocytes alone significantly impacts the activity of the TNR Notch reporter in the ovary.
Figure 9.
TNR (EGFP) expression in the ovaries of oocyte-specific Jag1 knockout mice. (A) Histology of WT and knockout (J1KO) ovaries at PND10, showing the presence of oocytes in the knockout ovaries. Scale bars, 100 µm. (B) Gene expression analysis indicating levels of Jag1 and Egfp mRNAs in normal littermates (NLM) (open bars) or cJ1KO/TNR (filled bars) ovaries at PND10. n = 4, with each animal shown as a point in the scatter plots. (C) Gene expression analysis indicating levels of Hes1, Hey2, and Heyl mRNAs in NLM (open bars) or cJ1KO/TNR (filled bars) ovaries at PND10. n = 5, with each animal an individual point in the scatter plots. The bars designate mean ± SEM. *P < 0.05.
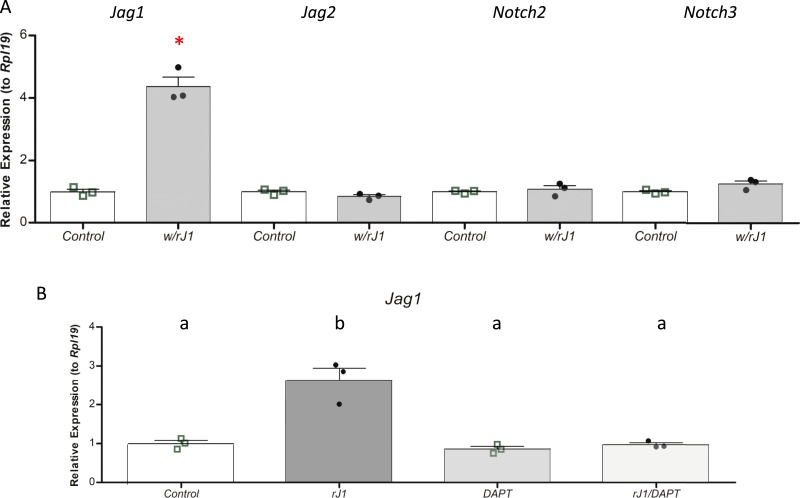
Finally, although JAG1 from the oocyte may activate Notch signaling in granulosa cells contacting the oocyte, or in those able to communicate with the oocyte through transzonal projections (39–41), there must be additional mechanisms that propagate and maintain this signal in growing multilayer follicles. In testing the activity of recombinant JAG1, we investigated its effects on the expression of genes encoding Notch ligands and receptors (Fig. 10). We observed that JAG1 stimulated its own expression while not altering expression of genes for the ligand Jag2 or the receptors Notch2 and Notch3 (Fig. 10A). The effect on Jag1 gene expression was specific and could be reserved with the pan-Notch inhibitor DAPT (Fig. 10B). This may provide a mechanism through which a Jag1-mediated Notch signal can be propagated across layers of granulosa cells within the growing follicle once activated in granulosa cells by the oocyte.
Figure 10.
Effects of recombinant JAG1 protein on Notch ligand and receptor gene expression in granulosa cells. Primary cultured granulosa cells were plated onto fibronectin (control) or immobilized recombinant Jag1 as described in the “Methods” section. (A) Expression of the Notch ligands Jag1 and Jag2 and the Notch receptors Notch2 and Notch3 was analyzed. (B) Expression of the ligand Jag1 was analyzed with or without a 24-h treatment with the Notch inhibitor DAPT. n = 3 for each condition, with individual experiments shown as points on the scatter plots. The bars designate mean ± SEM. In (A), *P < 0.05. In (B), means not sharing the same letter are significantly different (at P < 0.05).
Discussion
Communication between distinct ovarian cell types is critical for the formation, growth, and maturation of follicles as well as for regulating the developmental programs of these cells themselves. This is perhaps best understood in the context of the bidirectional signaling between the oocyte and surrounding somatic cells that will form the granulosa cells of the follicle. A variety of factors from the oocyte, including multiple TGF-β family proteins, act on granulosa cells to regulate their initial proliferation during follicle growth and their later acquisition of steroidogenic potential as they differentiate in mature follicles (42–45). Conversely, factors including both steroid and protein hormones from the granulosa cells impact the metabolism and growth of the oocyte as well as its eventual meiotic maturation (46–50). Although the role of secreted molecules in this intricately orchestrated and functionally important cellular cross-communication is well established, it has also been long recognized that direct physical contact between these juxtaposed cell types is also critical to follicle development. Gap junctions are one such form of physical communication (39, 40), and, indeed, disruption of the genes encoding the gap junction constituents Connexin-37 (Gja4) in the oocyte or Connexin-43 (Gja1) in granulosa cells results in female infertility (51–53).
In this study, we investigate another form of contact-dependent, or juxtacrine, cell communication via the Notch signaling pathway. The Notch pathway is one of the most highly conserved signaling systems in metazoan organisms, and it acts in a context-dependent fashion to regulate many aspects of cellular behavior during development (54–57). Its actions often involve cross-talk with various endocrine or paracrine signals (58–62). Recent studies using granulosa cell culture, ovary culture, and mouse knockout models have revealed Notch signaling to be important for the formation and growth of ovarian follicles and for female fertility (7–15). Consistent with these functions, numerous Notch ligands, receptors, and effector genes are expressed on the mouse ovary (9), and their abundance and distribution are developmentally regulated (7, 8, 10). Given this complexity, we have used a TNR mouse in which EGFP expression is regulated by the canonical Notch pathway obligate transcription factor Rbpj (16) to integrate and visualize Notch signaling activity in the mouse ovary (8). Using this reporter, we previously demonstrated that Notch activity is detected in the ovary by E15.5 in diffuse somatic cells near oocytes, an association that becomes increasingly apparent by birth during the early stages of primordial follicle formation (8).
To better understand the initial activation of Notch signaling in the ovary, we tested the hypothesis that signaling from the oocyte to surrounding somatic cells is necessary for Notch activation, by chemically or genetically ablating oocytes and assessing the impact on Notch signaling. The data demonstrate that reducing oocyte numbers during early development leads to a significant attenuation in the expression of Notch target genes and of the Notch activity reporter in the ovary, both in the late embryonic and early postnatal time periods. Furthermore, we show that the loss of the Notch ligand Jag1 from the oocyte can suppress Notch signaling in the ovary, whereas Jag1 alone is sufficient for activation of Notch signaling in granulosa cells. Collectively, these results support a role for the oocyte and Jag1 in the initial activation of Notch signaling in somatic granulosa cells at the time of follicle formation.
In both mouse models, the loss of oocytes and reduced expression of oocyte-specific markers was associated with significantly reduced levels of Notch target gene and reporter mRNAs, despite the relatively unchanged expression of several granulosa cell-specific genes. It is likely that this maintenance of granulosa cell identity is transitory, and will eventually be lost, as is the case in other mouse models of oocyte loss where granulosa cell transdifferentiation is observed (63–66). However, somatic cells in the white-spotted mouse ovary have previously been shown to normally express ovarian markers in the absence of oocytes during fetal life, arguing that germ cells are not required for the maturation of pregranulosa cells (28). Although TNR reporter expression was reduced in the two mouse models used in our studies, it was not eliminated, and remained at ∼30% of control levels. In busulfan-treated mice, this is likely in part explained by the substantial number of remaining oocytes that can be visually observed with the Sohlh1 and Vasa markers. A dose of busulfan that optimized oocyte loss while still allowing the embryos to survive was used for this study, but some oocytes do survive. Although many of the EGFP-expressing somatic cells are localized near these remaining oocytes, Notch-active cells can also be observed in areas devoid of oocytes. Because the timing of oocyte death in response to busulfan is likely variable, some of these TNR-positive cells may have been adjacent to viable oocytes before the oocytes themselves were lost. Consistent with our findings in the ovary, busulfan treatment in male mice to reduce testicular germ cell numbers also results in attenuated expression of the Notch target genes Hey1 and Hes1 (67), although residual expression of these genes remained.
Despite the effectiveness of the genetic white-spotted mouse model at reducing oocyte numbers, a basal level of TNR reporter expression remained in these ovaries as well. This may reflect activation of the reporter, and of Notch target genes, by noncanonical signaling mechanisms independent of the Rbpj transcription factor or of Notch ligand–receptor interactions (68–70). It also likely reflects Notch activity in ovarian cells other than granulosa cells. Using flow cytometry to sort both granulosa cells (Foxl2-positive) and Notch-active cells (TNR reporter), we found that ∼20% of the Notch-active cells in the PND21 ovary are not granulosa cells (36). Because Notch signaling is involved in vasculogenesis in many tissues (11, 14, 71–73), it is likely that this nongranulosa cell population includes vascular precursors such as endothelial cells in which Notch activity is expressed in response to stimuli other than the oocyte. In addition to the residual EGFP expression found diffusely throughout the ovary, strong expression was observed in granulosa cells of the rare follicles that formed around oocytes that had survived, consistent with these oocytes directly supporting Notch reporter gene activation.
Given the well-established cross-talk between Notch and a diversity of other signaling pathways (58–62), as well as the importance of the oocyte to the overall health of granulosa cells, many factors from the oocyte might contribute to regulating Notch activity in granulosa cells. However, the direct stimulus seemed likely to be an activating Notch ligand working in trans (74). The ligands Jag1 and Jag2 are abundantly expressed in the ovary (8, 9), whereas the three delta-like ligands are found at low levels at these stages but are induced by gonadotropin stimulation and have been implicated in later vascularization (11, 72, 73). Jag1 is predominantly found in the oocyte of early stage follicles (7–10) but is expressed in the granulosa cells of growing follicles and is strongly upregulated by gonadotropins in the periovulatory period (29). In contrast, Jag2 has been reported to be found in the oocyte (18) but it is more abundantly expressed in the granulosa cells of growing follicles (9). This suggested that Jag1 was the more likely candidate for an oocyte factor activating Notch signaling, and both the loss-of-function conditional knockout mice and the gain-of-function activation of Notch with recombinant Jag1 experiments reported here support this concept. A similar observation of Notch target gene activation using recombinant Jag1 ligand has been made in cultured testicular Sertoli cells (67). It remains possible that Jag2 from the oocyte is also involved, as we have demonstrated that recombinant Jag2 can activate Notch target genes in a manner similar to that reported here for recombinant Jag1 (36).
The processes by which a Notch signal might be propagated across multiple cell layers in a growing follicle remain poorly understood. In early stage follicles, granulosa cells not directly contacting the oocyte can remain in physical communication with it through specialized cytonemes termed transzonal projections, which are capable of traversing the zona pellucida and forming contacts to the oocyte at gap or adherens junctions (39–41). An attractive mechanism for signal propagation beyond this follicular stage is lateral induction, an established phenomenon in Notch signaling whereby a receiving cell upregulates expression of a ligand that activates receptors in adjacent cells allowing a wave of cells to be induced toward the same fate (75). Our finding that recombinant Jag1 induces its own expression in cultured granulosa cells, and the observation that Jag1 expression in granulosa cell increases as follicles mature, is consistent with Jag1 from the oocyte initiating an expansion of Notch activation across the follicle. Indeed, the TNR is highly expressed in larger preantral and antral follicles where mural granulosa cells have no contact with the oocyte (Fig. 7). Interestingly, recombinant Jag2 does not stimulate the expression of Jag1 (or of Jag2) in these cells (36), consistent with the notion that Jag1 is the more likely candidate for such a lateral induction mechanism.
Taken together, these data reveal an important role for the oocyte in initial activation of Notch signaling in granulosa cells during follicle formation. They support a model in which Notch ligands such as Jag1 from the oocyte act on Notch receptors, predominantly Notch2 and Notch3, on granulosa cells to stimulate the expression of Notch target genes, especially those of the Hey family of transcriptional repressors. The functional importance of these events and of Notch activation in granulosa cells is supported by the follicular phenotypes of mice lacking Notch2 (8, 15) or Hes1 (76) in granulosa cells, and by extensive data from ovary and granulosa cell cultures using Notch inhibitors, RNA interference knockdown, and neutralizing antibodies to reveal profound effects on granulosa cell survival and proliferation (7, 10, 11, 13, 20, 21, 28, 36). Numerous questions for future study remain. An important aspect of Notch signaling is the endocytosis and processing of both ligand in the sending cell and receptor in the receiving cell (77, 78), likely contributing to the bidirectional nature of the oocyte–granulosa cell communication. Much work remains to establish whether Jag1 does indeed mediate classical lateral induction (75) within the growing follicle and whether coexpression of ligands and receptors in granulosa cells leads to a related cis-inhibition phenomenon (74). Finally, further investigations to better appreciate the context-dependent nature of Notch signaling (79) as applied to the ovary and the regulation of female fertility are warranted.
Acknowledgments
Financial Support: This work was supported by Grant P01 HD021921 (to K.E.M.) from the National Institutes of Health Eunice Kennedy Shriver National Institute of Child Health and Human Development, and by training program Grant T32 GM008061 (to R.D.P.) from the National Institutes of Health Institute of General Medical Sciences.
Glossary
Abbreviations:
- cJ1KO
conditional Jagged1 knockout
- cN2KO
conditional Notch2 knockout
- DAPT
N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester
- E
embryonic day
- PND
postnatal day
- qRT-PCR
quantitative reverse transcription PCR
- S1CF
Sohlh1-mCherry
- TNR
transgenic Notch reporter
- WT
wild-type
Additional Information
Disclosure Summary: The authors have nothing to disclose.
Data Availability: All data generated or analyzed during this study are included in this published article or in the data repositories listed in References.
References and Notes
- 1. Richards JS, Pangas SA. The ovary: basic biology and clinical implications. J Clin Invest. 2010;120(4):963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barnett KR, Schilling C, Greenfeld CR, Tomic D, Flaws JA. Ovarian follicle development and transgenic mouse models. Hum Reprod Update. 2006;12(5):537–555. [DOI] [PubMed] [Google Scholar]
- 3. Kerr JB, Myers M, Anderson RA. The dynamics of the primordial follicle reserve. Reproduction. 2013;146(6):R205–R215. [DOI] [PubMed] [Google Scholar]
- 4. Matzuk MM, Burns KH, Viveiros MM, Eppig JJ. Intercellular communication in the mammalian ovary: oocytes carry the conversation. Science. 2002;296(5576):2178–2180. [DOI] [PubMed] [Google Scholar]
- 5. Eppig JJ. Reproduction: oocytes call, granulosa cells connect. Curr Biol. 2018;28(8):R354–R356. [DOI] [PubMed] [Google Scholar]
- 6. Schultz RM. Roles of cell-to-cell communication in development. Biol Reprod. 1985;32(1):27–42. [DOI] [PubMed] [Google Scholar]
- 7. Vanorny DA, Mayo KE. The role of Notch signaling in the mammalian ovary. Reproduction. 2017;153(6):R187–R204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vanorny DA, Prasasya RD, Chalpe AJ, Kilen SM, Mayo KE. Notch signaling regulates ovarian follicle formation and coordinates follicular growth. Mol Endocrinol. 2014;28(4):499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Johnson J, Espinoza T, McGaughey RW, Rawls A, Wilson-Rawls J. Notch pathway genes are expressed in mammalian ovarian follicles. Mech Dev. 2001;109(2):355–361. [DOI] [PubMed] [Google Scholar]
- 10. Trombly DJ, Woodruff TK, Mayo KE. Suppression of Notch signaling in the neonatal mouse ovary decreases primordial follicle formation. Endocrinology. 2009;150(2):1014–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jovanovic VP, Sauer CM, Shawber CJ, Gomez R, Wang X, Sauer MV, Kitajewski J, Zimmermann RC. Intraovarian regulation of gonadotropin-dependent folliculogenesis depends on notch receptor signaling pathways not involving Delta-like ligand 4 (Dll4). Reprod Biol Endocrinol. 2013;11(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hahn KL, Johnson J, Beres BJ, Howard S, Wilson-Rawls J. Lunatic fringe null female mice are infertile due to defects in meiotic maturation. Development. 2005;132(4):817–828. [DOI] [PubMed] [Google Scholar]
- 13. Zhang CP, Yang JL, Zhang J, Li L, Huang L, Ji SY, Hu ZY, Gao F, Liu YX. Notch signaling is involved in ovarian follicle development by regulating granulosa cell proliferation. Endocrinology. 2011;152(6):2437–2447. [DOI] [PubMed] [Google Scholar]
- 14. Vorontchikhina MA, Zimmermann RC, Shawber CJ, Tang H, Kitajewski J. Unique patterns of Notch1, Notch4 and Jagged1 expression in ovarian vessels during folliculogenesis and corpus luteum formation. Gene Expr Patterns. 2005;5(5):701–709. [DOI] [PubMed] [Google Scholar]
- 15. Xu J, Gridley T. Notch2 is required in somatic cells for breakdown of ovarian germ-cell nests and formation of primordial follicles. BMC Biol. 2013;11(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duncan AW, Rattis FM, DiMascio LN, Congdon KL, Pazianos G, Zhao C, Yoon K, Cook JM, Willert K, Gaiano N, Reya T. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat Immunol. 2005;6(3):314–322. [DOI] [PubMed] [Google Scholar]
- 17. Murta D, Batista M, Silva E, Trindade A, Mateus L, Duarte A, Lopes-da-Costa L. Differential expression of Notch component and effector genes during ovarian follicle and corpus luteum development during the oestrous cycle. Reprod Fertil Dev. 2015;27(7):1038–1048. [DOI] [PubMed] [Google Scholar]
- 18. Guo M, Zhang H, Bian F, Li G, Mu X, Wen J, Mao G, Teng Z, Xia G, Zhang M. P4 down-regulates Jagged2 and Notch1 expression during primordial folliculogenesis. Front Biosci (Elite Ed). 2012;4:2631–2644. [DOI] [PubMed] [Google Scholar]
- 19. Dovey HF, John V, Anderson JP, Chen LZ, de Saint Andrieu P, Fang LY, Freedman SB, Folmer B, Goldbach E, Holsztynska EJ, Hu KL, Johnson-Wood KL, Kennedy SL, Kholodenko D, Knops JE, Latimer LH, Lee M, Liao Z, Lieberburg IM, Motter RN, Mutter LC, Nietz J, Quinn KP, Sacchi KL, Seubert PA, Shopp GM, Thorsett ED, Tung JS, Wu J, Yang S, Yin CT, Schenk DB, May PC, Altstiel LD, Bender MH, Boggs LN, Britton TC, Clemens JC, Czilli DL, Dieckman-McGinty DK, Droste JJ, Fuson KS, Gitter BD, Hyslop PA, Johnstone EM, Li WY, Little SP, Mabry TE, Miller FD, Audia JE. Functional gamma-secretase inhibitors reduce beta-amyloid peptide levels in brain. J Neurochem. 2001;76(1):173–181. [DOI] [PubMed] [Google Scholar]
- 20. Chen CL, Fu XF, Wang LQ, Wang JJ, Ma HG, Cheng SF, Hou ZM, Ma JM, Quan GB, Shen W, Li L. Primordial follicle assembly was regulated by Notch signaling pathway in the mice. Mol Biol Rep. 2014;41(3):1891–1899. [DOI] [PubMed] [Google Scholar]
- 21. Terauchi KJ, Shigeta Y, Iguchi T, Sato T. Role of Notch signaling in granulosa cell proliferation and polyovular follicle induction during folliculogenesis in mouse ovary. Cell Tissue Res. 2016;365(1):197–208. [DOI] [PubMed] [Google Scholar]
- 22. Bucci LR, Meistrich ML. Effects of busulfan on murine spermatogenesis: cytotoxicity, sterility, sperm abnormalities, and dominant lethal mutations. Mutat Res. 1987;176(2):259–268. [DOI] [PubMed] [Google Scholar]
- 23. Hemsworth BN, Jackson H. Effect of busulphan on the developing ovary in the rat. J Reprod Fertil. 1963;6(2):229–233. [DOI] [PubMed] [Google Scholar]
- 24. Suzuki H, Dann CT, Rajkovic A. Generation of a germ cell-specific mouse transgenic CHERRY reporter, Sohlh1-mCherryFlag. Genesis. 2013;51(1):50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Little CC, Cloudman AM. The occurrence of a dominant spotting mutation in the house mouse. Proc Natl Acad Sci USA. 1937;23(10):535–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smith ER, Yeasky T, Wei JQ, Miki RA, Cai KQ, Smedberg JL, Yang WL, Xu XX. White spotting variant mouse as an experimental model for ovarian aging and menopausal biology. Menopause. 2012;19(5):588–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hubbard N, Prasasya RD, Mayo KE. Data from: Activation of Notch signaling by oocytes and Jag1 in mouse ovarian granulosa cells. figshare 2019. Deposited 2 October 2019. 10.6084/m9.figshare.9875939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maatouk DM, Mork L, Hinson A, Kobayashi A, McMahon AP, Capel B. Germ cells are not required to establish the female pathway in mouse fetal gonads. PLoS One. 2012;7(10):e47238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Prasasya RD, Mayo KE. Notch signaling regulates differentiation and steroidogenesis in female mouse ovarian granulosa cells. Endocrinology. 2018;159(1):184–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3(6):1101–1108. [DOI] [PubMed] [Google Scholar]
- 31. Park OK, Mayo KE. Transient expression of progesterone receptor messenger RNA in ovarian granulosa cells after the preovulatory luteinizing hormone surge. Mol Endocrinol. 1991;5(7):967–978. [DOI] [PubMed] [Google Scholar]
- 32. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. RRID:AB_443012, https://scicrunch.org/resolver/AB_443012.
- 34. Pangas SA, Choi Y, Ballow DJ, Zhao Y, Westphal H, Matzuk MM, Rajkovic A. Oogenesis requires germ cell-specific transcriptional regulators Sohlh1 and Lhx8. Proc Natl Acad Sci USA. 2006;103(21):8090–8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. RRID:AB_221569, https://scicrunch.org/resolver/AB_221569.
- 36. Prasasya RD. Regulation of Ovarian Granulosa Cell Proliferation and Differentiation by the Notch Signaling Pathway [dissertation]. Evanston, IL: Northwestern University; 2018. [Google Scholar]
- 37. Garcia TX, Farmaha JK, Kow S, Hofmann MC. RBPJ in mouse Sertoli cells is required for proper regulation of the testis stem cell niche. Development. 2014;141(23):4468–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dong Y, Long T, Wang C, Mirando AJ, Chen J, O’Keefe RJ, Hilton MJ. NOTCH-mediated maintenance and expansion of human bone marrow stromal/stem cells: a technology designed for orthopedic regenerative medicine. Stem Cells Transl Med. 2014;3(12):1456–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mora JM, Fenwick MA, Castle L, Baithun M, Ryder TA, Mobberley M, Carzaniga R, Franks S, Hardy K.. Characterization and significance of adhesion and junction-related proteins in mouse ovarian follicles. Biol Reprod. 2012;86(5):153, 1–14. doi: 10.1095/biolreprod.111.096156. [DOI] [PubMed] [Google Scholar]
- 40. Clarke HJ. Regulation of germ cell development by intercellular signaling in the mammalian ovarian follicle. Wiley Interdiscip Rev Dev Biol. 2018;7(1):e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Baena V, Terasaki M. Three-dimensional organization of transzonal projections and other cytoplasmic extensions in the mouse ovarian follicle. Sci Rep. 2019;9(1):1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yan C, Wang P, DeMayo J, DeMayo FJ, Elvin JA, Carino C, Prasad SV, Skinner SS, Dunbar BS, Dube JL, Celeste AJ, Matzuk MM. Synergistic roles of bone morphogenetic protein 15 and growth differentiation factor 9 in ovarian function. Mol Endocrinol. 2001;15(6):854–866. [DOI] [PubMed] [Google Scholar]
- 43. Dong J, Albertini DF, Nishimori K, Kumar TR, Lu N, Matzuk MM. Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature. 1996;383(6600):531–535. [DOI] [PubMed] [Google Scholar]
- 44. Rimon-Dahari N, Yerushalmi-Heinemann L, Alyagor L, Dekel N. Ovarian folliculogenesis. Results Probl Cell Differ. 2016;58:167–190. [DOI] [PubMed] [Google Scholar]
- 45. Trombly DJ, Woodruff TK, Mayo KE. Roles for transforming growth factor beta superfamily proteins in early folliculogenesis. Semin Reprod Med. 2009;27(1):14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kezele P, Skinner MK. Regulation of ovarian primordial follicle assembly and development by estrogen and progesterone: endocrine model of follicle assembly. Endocrinology. 2003;144(8):3329–3337. [DOI] [PubMed] [Google Scholar]
- 47. Chen Y, Jefferson WN, Newbold RR, Padilla-Banks E, Pepling ME. Estradiol, progesterone, and genistein inhibit oocyte nest breakdown and primordial follicle assembly in the neonatal mouse ovary in vitro and in vivo. Endocrinology. 2007;148(8):3580–3590. [DOI] [PubMed] [Google Scholar]
- 48. Myers M, Middlebrook BS, Matzuk MM, Pangas SA. Loss of inhibin alpha uncouples oocyte-granulosa cell dynamics and disrupts postnatal folliculogenesis. Dev Biol. 2009;334(2):458–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Namwanje M, Brown CW. Activins and inhibins: roles in development, physiology, and disease. Cold Spring Harb Perspect Biol. 2016;8(7):a021881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang M, Su YQ, Sugiura K, Wigglesworth K, Xia G, Eppig JJ. Estradiol promotes and maintains cumulus cell expression of natriuretic peptide receptor 2 (NPR2) and meiotic arrest in mouse oocytes in vitro. Endocrinology. 2011;152(11):4377–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Simon AM, Chen H, Jackson CL. Cx37 and Cx43 localize to zona pellucida in mouse ovarian follicles. Cell Commun Adhes. 2006;13(1–2):61–77. [DOI] [PubMed] [Google Scholar]
- 52. Gittens JE, Kidder GM. Differential contributions of connexin37 and connexin43 to oogenesis revealed in chimeric reaggregated mouse ovaries. J Cell Sci. 2005;118(Pt 21):5071–5078. [DOI] [PubMed] [Google Scholar]
- 53. Li TY, Colley D, Barr KJ, Yee SP, Kidder GM. Rescue of oogenesis in Cx37-null mutant mice by oocyte-specific replacement with Cx43. J Cell Sci. 2007;120(Pt 23):4117–4125. [DOI] [PubMed] [Google Scholar]
- 54. Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138(17):3593–3612. [DOI] [PubMed] [Google Scholar]
- 55. Henrique D, Schweisguth F. Mechanisms of Notch signaling: a simple logic deployed in time and space. Development. 2019;146(3):dev172148. [DOI] [PubMed] [Google Scholar]
- 56. Siebel C, Lendahl U. Notch signaling in development, tissue homeostasis, and disease. Physiol Rev. 2017;97(4):1235–1294. [DOI] [PubMed] [Google Scholar]
- 57. Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hurlbut GD, Kankel MW, Lake RJ, Artavanis-Tsakonas S. Crossing paths with Notch in the hyper-network. Curr Opin Cell Biol. 2007;19(2):166–175. [DOI] [PubMed] [Google Scholar]
- 59. Dahlqvist C, Blokzijl A, Chapman G, Falk A, Dannaeus K, Ibâñez CF, Lendahl U. Functional Notch signaling is required for BMP4-induced inhibition of myogenic differentiation. Development. 2003;130(24):6089–6099. [DOI] [PubMed] [Google Scholar]
- 60. Itoh F, Itoh S, Goumans MJ, Valdimarsdottir G, Iso T, Dotto GP, Hamamori Y, Kedes L, Kato M, ten Dijke Pt P. Synergy and antagonism between Notch and BMP receptor signaling pathways in endothelial cells. EMBO J. 2004;23(3):541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang LQ, Liu JC, Chen CL, Cheng SF, Sun XF, Zhao Y, Yin S, Hou ZM, Pan B, Ding C, Shen W, Zhang XF. Regulation of primordial follicle recruitment by cross-talk between the Notch and phosphatase and tensin homologue (PTEN)/AKT pathways. Reprod Fertil Dev. 2016;28(6):700–712. [DOI] [PubMed] [Google Scholar]
- 62. Dorfman MD, Kerr B, Garcia-Rudaz C, Paredes AH, Dissen GA, Ojeda SR. Neurotrophins acting via TRKB receptors activate the JAGGED1-NOTCH2 cell-cell communication pathway to facilitate early ovarian development. Endocrinology. 2011;152(12):5005–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Merchant H. Rat gonadal and ovarian organogenesis with and without germ cells. An ultrastructural study. Dev Biol. 1975;44(1):1–21. [DOI] [PubMed] [Google Scholar]
- 64. Merchant-Larios H, Taketo T. Testicular differentiation in mammals under normal and experimental conditions. J Electron Microsc Tech. 1991;19(2):158–171. [DOI] [PubMed] [Google Scholar]
- 65. Guigon CJ, Magre S. Contribution of germ cells to the differentiation and maturation of the ovary: insights from models of germ cell depletion. Biol Reprod. 2006;74(3):450–458. [DOI] [PubMed] [Google Scholar]
- 66. Merchant-Larios H, Centeno B. Morphogenesis of the ovary from the sterile W/Wv mouse. Prog Clin Biol Res. 1981;59B:383–392. [PubMed] [Google Scholar]
- 67. Garcia TX, Parekh P, Gandhi P, Sinha K, Hofmann MC. The NOTCH ligand JAG1 regulates GDNF expression in Sertoli cells. Stem Cells Dev. 2017;26(8):585–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Heitzler P. Biodiversity and noncanonical Notch signaling. Curr Top Dev Biol. 2010;92:457–481. [DOI] [PubMed] [Google Scholar]
- 69. D’Souza B, Meloty-Kapella L, Weinmaster G. Canonical and non-canonical Notch ligands. Curr Top Dev Biol. 2010;92:73–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Andersen P, Uosaki H, Shenje LT, Kwon C. Non-canonical Notch signaling: emerging role and mechanism. Trends Cell Biol. 2012;22(5):257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gridley T. Notch signaling in the vasculature. Curr Top Dev Biol. 2010;92:277–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Fraser HM, Hastings JM, Allan D, Morris KD, Rudge JS, Wiegand SJ. Inhibition of delta-like ligand 4 induces luteal hypervascularization followed by functional and structural luteolysis in the primate ovary. Endocrinology. 2012;153(4):1972–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Xie Q, Cheng Z, Chen X, Lobe CG, Liu J. The role of Notch signalling in ovarian angiogenesis. J Ovarian Res. 2017;10(1):13–017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. del Álamo D, Rouault H, Schweisguth F. Mechanism and significance of cis-inhibition in Notch signalling. Curr Biol. 2011;21(1):R40–R47. [DOI] [PubMed] [Google Scholar]
- 75. Sjöqvist M, Andersson ER. Do as I say, Not(ch) as I do: lateral control of cell fate. Dev Biol. 2019;447(1):58–70. [DOI] [PubMed] [Google Scholar]
- 76. Manosalva I, González A, Kageyama R. Hes1 in the somatic cells of the murine ovary is necessary for oocyte survival and maturation. Dev Biol. 2013;375(2):140–151. [DOI] [PubMed] [Google Scholar]
- 77. Musse AA, Meloty-Kapella L, Weinmaster G. Notch ligand endocytosis: mechanistic basis of signaling activity. Semin Cell Dev Biol. 2012;23(4):429–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kandachar V, Roegiers F. Endocytosis and control of Notch signaling. Curr Opin Cell Biol. 2012;24(4):534–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bray SJ. Notch signalling in context. Nat Rev Mol Cell Biol. 2016;17(11):722–735. [DOI] [PubMed] [Google Scholar]