Figure 5.
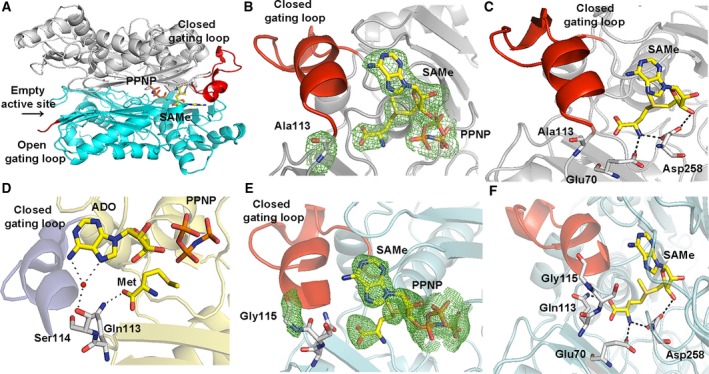
Comparison of Q113A and P115G MATα2 mutant and wild‐type structure. (A) Two different conformations of dimeric subunits of the enzyme are shown. One monomer has a closed gating loop (red) with SAMe and PPNP inside the active site. The other subunit has the disordered, open gating loop (red) and no ligands in the active site. (B) A close view of Q113A mutant shows the omit map contoured around Ala113 and products (PPNP and SAMe) in the active site pocket with ordered and closed gating loops (red). Fo‐Fc omit maps are contoured at the 3 σ level and colored in green. (C) A close view of SAMe in the active site of Q113A mutant. (D) The interactions of Gln113 and Ser114 with Met and ADO in wt MATα2. Water is shown as a red sphere. (E) The close view of P115G mutant shows well‐ordered, closed gating loops and the Fo‐Fc omit map is colored in green and contoured at the 3 σ level around Gly115 and products (SAMe, PPNP). (F) A close view of SAMe in the active site of P115G mutant.
