Figure 3.
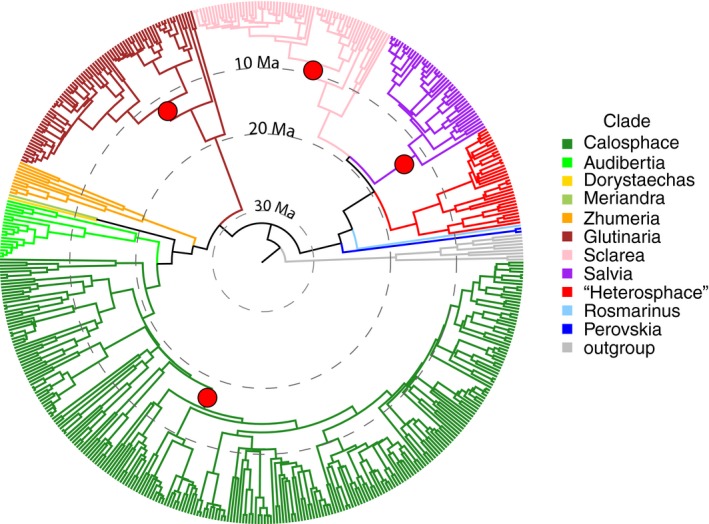
BEAST maximum clade credibility tree (yule model) using Salvia nrDNA and rooted with a monophyletic outgroup of Lepechinia and Melissa. Major clades that were well supported based on AHE genomic data (see Fig. 2) were used as topological constraints in BEAST. Edges are colored by subgenus. (The informal “Heterosphace” is used here and includes New World “Salviastrum” and Old World “Verticillata” and their relatives; the “Salvia aegyptiaca clade” is informally placed here with previously recognized subgen. Zhumeria). The edges with the four BAMM species diversification shifts (see Fig. 4) are indicated. This MCC tree is used for all other subsequent analyses. An enlarged BEAST tree with species terminals and 95% nodal age intervals is provided in Appendix S8.
