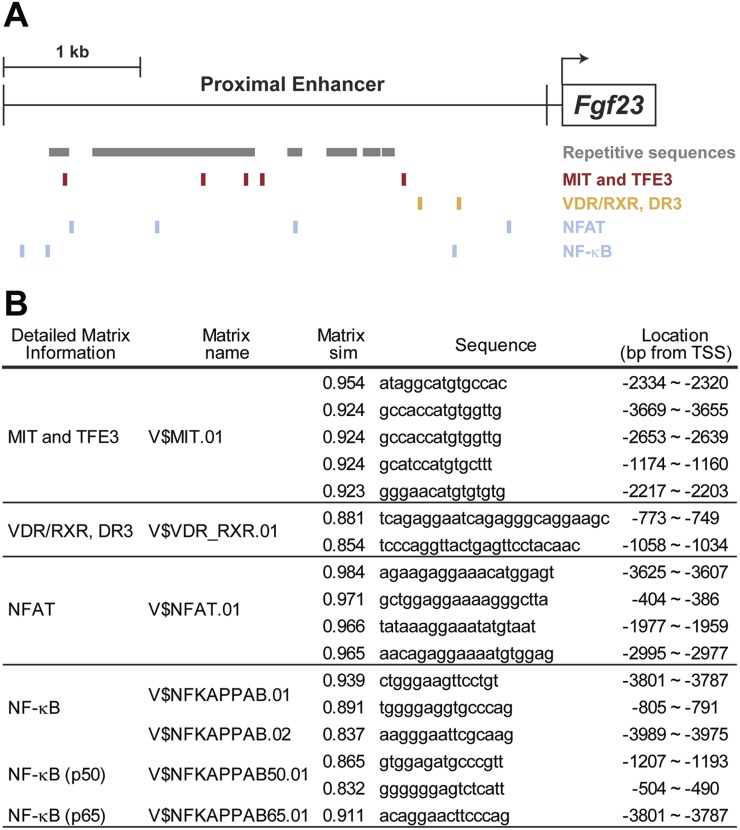
Figure 7.
Putative transcription factor binding motifs in proximal enhancer region. (A) Repetitive sequences defined by the University of California, Santa Cruz Genome Browser (genome.uscs.edu) are marked in the proximal regulatory region as gray boxes. Locations of potential binding motifs of MITF/TFE3, VDR/RXR, NFAT, and NF-κB are presented. Transcription start site is shown by an arrow. (B) Putative transcription factor binding motifs were predicted by MatInspector as described in the “Materials and Methods.” Matrix names, matrix similarity (Matrix sim), sequences, and locations of the motifs for MITF/TFE3, VDR/RXR, NFAT, and NF-κB are presented. MIT, microphthalmia transcription factor; RXR, retinoid X receptor; TSS, transcription start site.