Figure 2.
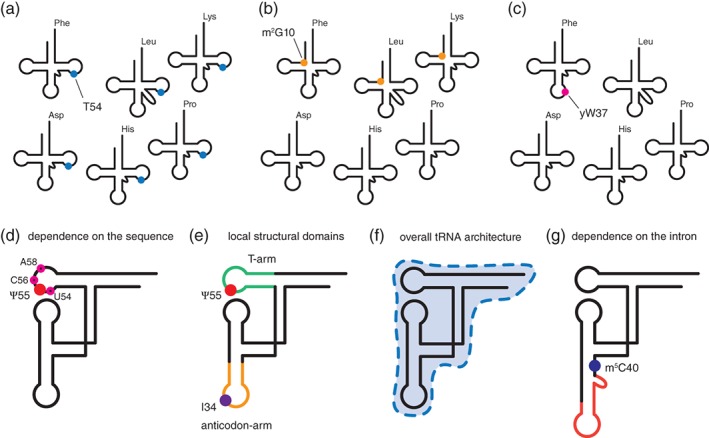
Determinants of specificity in tRNA modification enzymes. (a–c) Modification enzymes can be classified into three classes depending on their degree of specificity with respect to tRNA identities. Examples are drawn based on the specificity found in the yeast S. cerevisiae. (a) The first class consists of modification enzymes that modify all tRNAs with the correct target nucleotide, such as Trm2 introducing T54 in all cytoplasmic tRNAs in yeast. (b) The second class consists of modification enzymes that modify only a subset of tRNAs having the correct target nucleotide, such as Trm11/Trm112 introducing m2G10 in approximately half of the tRNAs in yeast. (c) The third class consists of modification enzymes that modify only a single tRNA species. For instance in yeast, the complex modification wybutosine (yW) is found uniquely at position 37 of tRNAPhe. (d–g) General principles controlling substrate recognition by tRNA modification enzymes. These general principles include (d) dependence on the RNA sequence, (e) dependence on local structural domains, (f) dependence on the overall tRNA architecture, and (g) dependence on the presence of an intronic sequence. A combination of these different principles determines the actual specificity of tRNA modification enzymes, which often involves both a certain degree of sequence specificity and structural determinants.
