Figure 5.
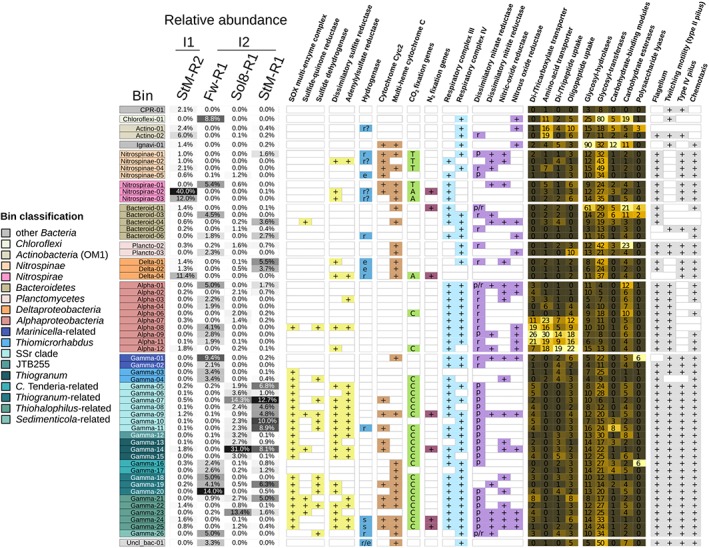
Metabolic potential of abundant microbial populations. Relative abundance of bins (>1%) in the metagenomes is shown in the first four columns of the figure. The presence or absence of a pathway/enzyme is based on presence of essential key enzymes/catalytic subunits. Hydrogenases were classified according to HydDB (Søndergaard et al., 2016) into ‘r’– respiratory/H2‐oxidizing, ‘r?’– unknown, probably H2‐oxidizing, reactive oxygen species‐protecting, ‘e’ – H2‐evolving and ‘s’ – H2‐sensing. For the carbon fixation pathways ‘A’ indicates acetyl‐CoA pathway, ‘R’ – Calvin–Benson–Bassham cycle, ‘T’–reverse TCA cycle. Nitrate reductases are denoted as ‘p’–periplasmic and ‘r’–respiratory/membrane bound. For genes expected to be found in most living organisms, numbers and a heatmap are used to indicate the amount of genes found in a given bin.
