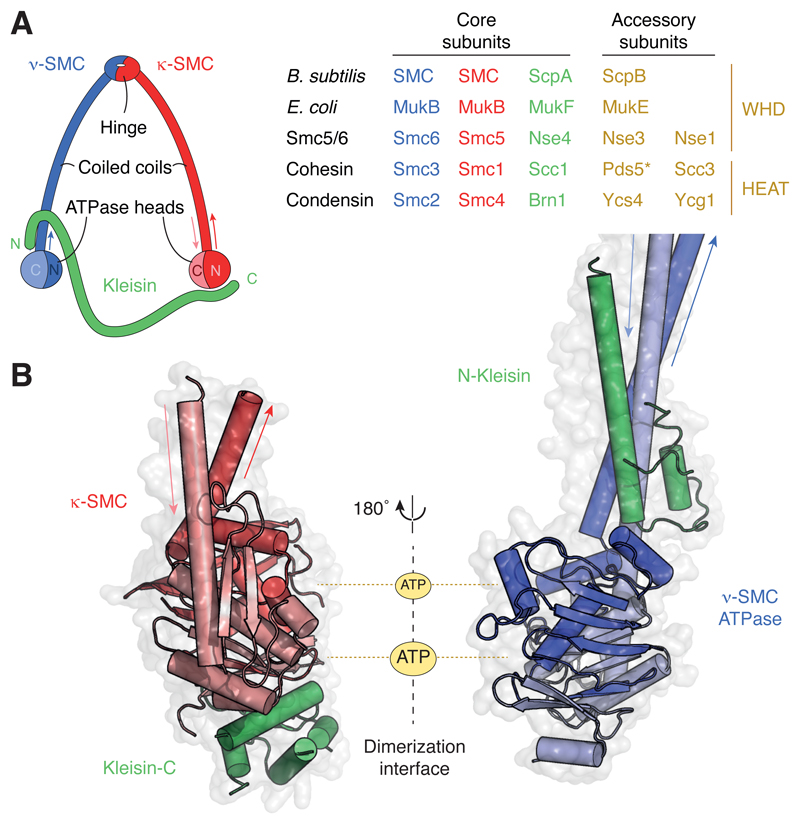
Figure 1. Architecture and composition of SMC complexes.
(A) Ring-like architecture created by the association of the SMC and kleisin subunits and subunit composition of common prokaryotic (top two) and eukaryotic (bottom three; S. cerevisiae protein names) SMC complexes. *Note that Pds5 associates with the other cohesin subunits transiently and might hence not be a stoichiometric subunit of cohesin complexes. (B) Structure models of a κ-SMC ATPase head domain bound to the WHD located at the C terminus of the kleisin subunit (pdb: 1w1w; S. cerevisiae cohesin Smc1–Scc1) and of a ν-SMC ATPase head domain bound to the helical domain located at the N terminus of the kleisin subunit (pdb: 4ux3; S. cerevisiae cohesin Smc3–Scc1).