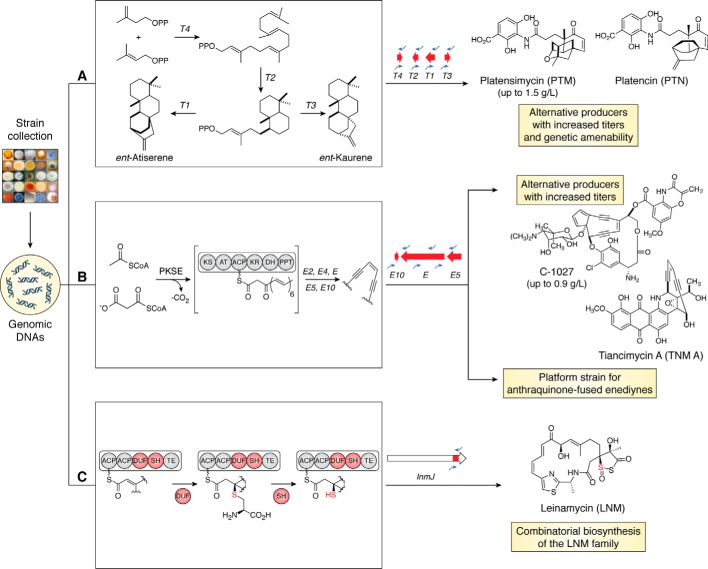
Figure 3.
Examples of structure-centric approaches to leverage the microbial strain collection at TSRI for natural product discovery. A, PTM and PTN contain unique diterpenoid scaffolds, the biosynthesis of which is encoded by terpene cyclases, T1, T2, T3, and T4. An RT-PCR screen of a fraction of the actinobacteria strains in the collection prioritized strains containing all four cyclase genes, resulting in the identification of alternative PTM and PTN producers with high titers and superior growth characteristics. B, enediyne natural products contain a unique pharmacophore, consisting of two triple bonds in conjugation with a double bond or incipient double bond, the biosynthesis of which is encoded by the enediyne PKS gene cassette, E, E10, E3, E4, and E5. An RT-PCR screen of a fraction of the actinobacterial strains in the collection prioritized strains containing the enediyne PKS gene cassette, resulting in the identification of alternative C-1027 producers with high titers and new anthraquinone-fused enediynes TNMs. The TNM producer, with its high TNM titers and superior growth characteristics, has enabled the development of a platform strain for engineered biosynthesis and production of the anthraquinone-fused family of enediyne natural products. C, LNM contains a unique sulfur-containing heterocycle, where the sulfur incorporation chemistry is encoded by a DUF-SH didomain. An RT-PCR screen of a fraction of the actinobacterial strains in the collection prioritized strains containing the DUF-SH didomain, resulting in the identification of a family of modular biosynthetic pathways that exemplify how Nature does combinatorial biosynthesis for the LNM family of natural products.