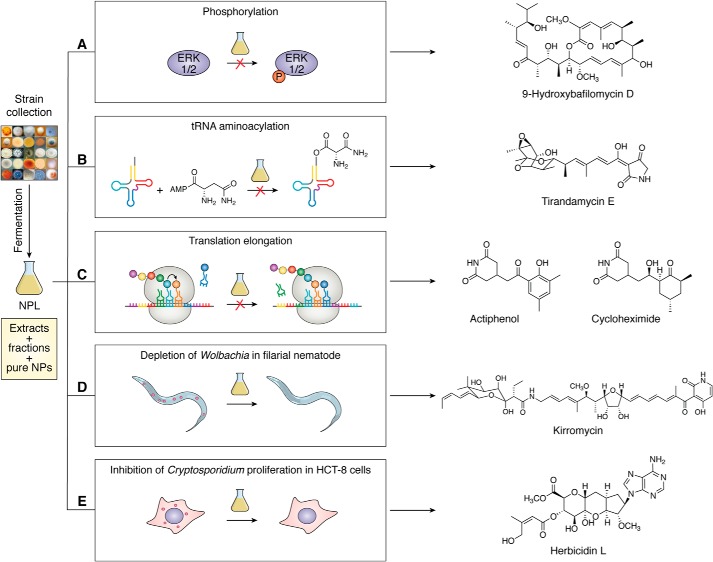
Figure 4.
Examples of function-centric approaches to leverage the microbial strain collection at TSRI for natural product discovery. Selected strains from the collection are fermented in diverse media to make the crude extracts, and upon HPLC analysis, the most chemically diverse conditions are selected, scaled up, and subjected to chromatography to afford the partially purified fractions. The current NPL consists of crude extracts, partially purified fractions, and pure natural products (also see Fig. 1B) and has been subjected to HTS, against emerging biology, for natural product discovery. A, NPL was screened for cytotoxicity and prolactin-initiated phosphorylation of ERK1/2. Bioassay-guided dereplication of the active hits led to the isolation of two new bafilomycin congeners, along with nine known ones. B, NPL was screened for parasitic AsnRS inhibition. Bioassay-guided dereplication of the hits identified TAMs as potent and selective AsnRS inhibitors, three of which were new congeners, and two were previously characterized. C, NPL was screened for inhibition of protein translation initiation via a high-throughput bicistronic mRNA translation assay. Bioassay-guided dereplication of the hits led to the identification of actiphenol and cycloheximide as potent inhibitors, importantly, from the same Streptomyces strain, shedding new insights into their biosynthesis. D, pure natural product collection of the NPL was subjected to a high-content image-based screening to discover inhibitors of Wolbachia via a Drosophila infection model, resulting in the identification of kirromycin as a potent and specific inhibitor of Wolbachia. E, partially-purified fraction collection of the NPL was subjected to a high-content image-based screen to search for inhibitors of Cryptosporidium via a JW18 infection model, leading to the discovery of the herbicidins as a promising scaffold for anti-Cryptosporidium drug development.