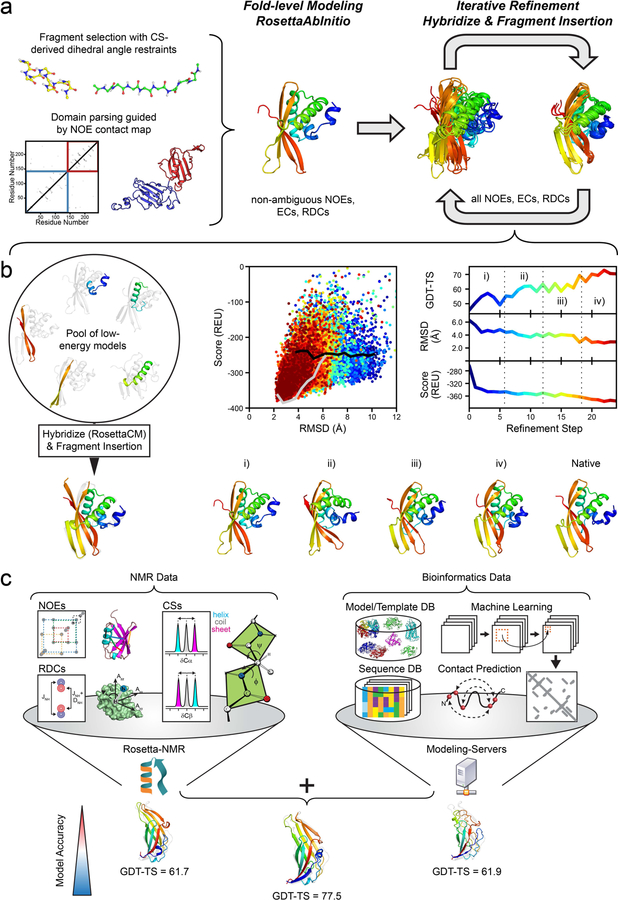
Figure 1. Overview of the modeling protocol employed in the CASP13 NMR-assisted structure prediction category.
(a) Overall flowchart: Initial fold-level modeling was done with RosettaAbInitio guided by non-ambiguous NOEs, evolutionary coupling restraints (ECs) and RDCs. The library of 3mer and 9mer peptide fragments was created with the Rosetta Fragment Picker using CS-derived ϕ/ψ dihedral angle restraints. Proteins with >200 residues were manually parsed into domains guided by non-ambiguous NOE contacts, and domains were modeled separately. After fold-level modeling, models were iteratively refined by hybridization and fragment insertion guided by all ambiguous and non-ambiguous NOEs, ECs and RDCs. (b) Improvement in model accuracy for target N0968s1 through an iterative refinement protocol. The protocol maintains a pool of low-energy models which are hybridized with RosettaCM and diversified through fragment insertion. The protocol stops when models are converged in terms of their pairwise GDT-HA or the final refinement step is reached. The bottom row demonstrates the improvement in the accuracy of N0968s1 models as the protocol proceeds: (i) GDT-TS = 47.5, (ii) GDT-TS = 58.0, (iii) GDT-TS = 60.2, (iv) GDT-TS = 72.4. (c) Suggested meta-approach to NMR structure modeling by incorporating structural restraints retrieved from bioinformatical sources/databases (DB), e.g. predicted residue-residue contacts and structural templates. In our post-CASP13 analysis, we used the initial predictions of five different servers (Robetta35, I-TASSER36,37, QUARK38, RaptorX-Contact39, RaptorX-TBM40) and refined them iteratively with Rosetta and NMR data leading to more accurate models than each individual technique. Model improvement is exemplified for target N0981-D5 for which the GDT-TS score increased by >15.