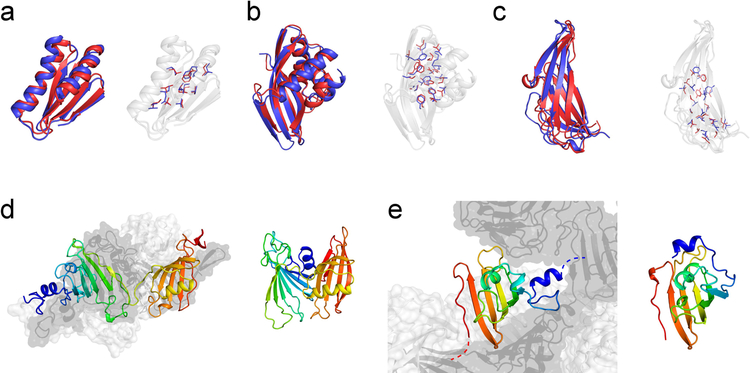
Figure 3. Examples of successful and difficult modeling cases.
High-resolution structure predictions could be made for three targets, (a) N1008 (Cα-RMSD = 2.06 Å), (b) N0968s1 (Cα-RMSD = 2.88 Å), and (c) N0981-D5 (Cα-RMSD = 3.48 Å), allowing accurate sidechain placement. The submitted Model 1 (red) is displayed as cartoon representation and compared with the experimental reference structure (blue). Sidechains in the protein core are depicted with sticks. The fraction of correct χ1 and χ2 rotamers in buried protein regions was (a) 74% / 37%, (b) 77% / 41%, and (c) 66% / 29%, respectively. Difficult modeling cases were proteins with quaternary structure which have interfaces to other protein copies or domains. (d) Left: experimental structure of the homo-trimeric target N0989. Chain A is shown as cartoon and is rainbow-colored. Chains B and C are shown in surface representation and colored light and dark gray, respectively. Right: The submitted model for the monomeric protein had a too compact, non-extended conformation. (e) Left: experimental structure of domain D4 of the homo-trimeric target N0981. The N-terminal helix (blue) is linked to another domain in the native assembly and stabilized by interactions with the two neighboring subunits indicated in light and dark gray, respectively. Right: The submitted model for N0981-D4 shows an incorrect orientation of the N-terminal helix because no NOE contact information was available to restrain this part of the structure.