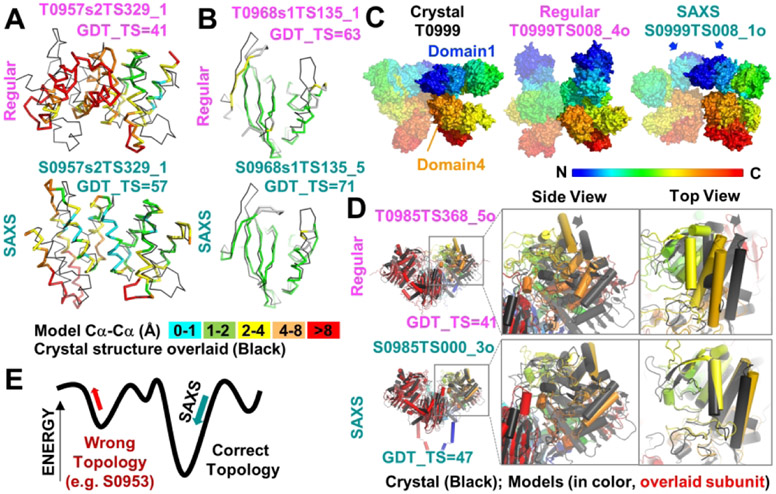
Figure 7.
Examples where SAXS-assisted models were better than best regular model from all groups or from the same group. (A and B) SAXS-assisted models for targets S0957s2 and S0968s1, respectively, had higher GDT_TS scores than regular models from the same group. Ribbon diagrams of domain models (colored by Cα-Cα deviation from the crystal) are overlaid onto the respective crystal structure (black). (C) The Pierce-group SAXS assisted assembly model for target S0999 was visually better than the best regular model from the same group. Surface models are colored by rainbow from the N to C terminus. One subunit of homodimer is partially transparent so that chains can be distinguished. Arrows highlight the domain 1 dimer interface that is predicted in the SAXS-assisted model. (D) The 3Dbio SAXS-assisted homodimer model for target S0985 had a better GDT_TS score for the entire ensemble than the best regular model from any group and better overlaid on the crystal structure. Arrows on regular model highlight rotation needed for proper overlay. Cartoon depiction with cylindrical helices of models when the left subunit (red) is overlaid onto crystal structure. The right subunit is colored by rainbow as in C and is the focus of the zoom views. (E) A model for how inclusion of SAXS data would have opposing effects on the fold energy term, depending on the starting model topology. If the starting model has the wrong topology, SAXS data would distort the wrong topology into the right shape. If the starting model has the right topology, SAXS data would lead to an improved fold with no deterioration of the folding elements.