Figure 3.
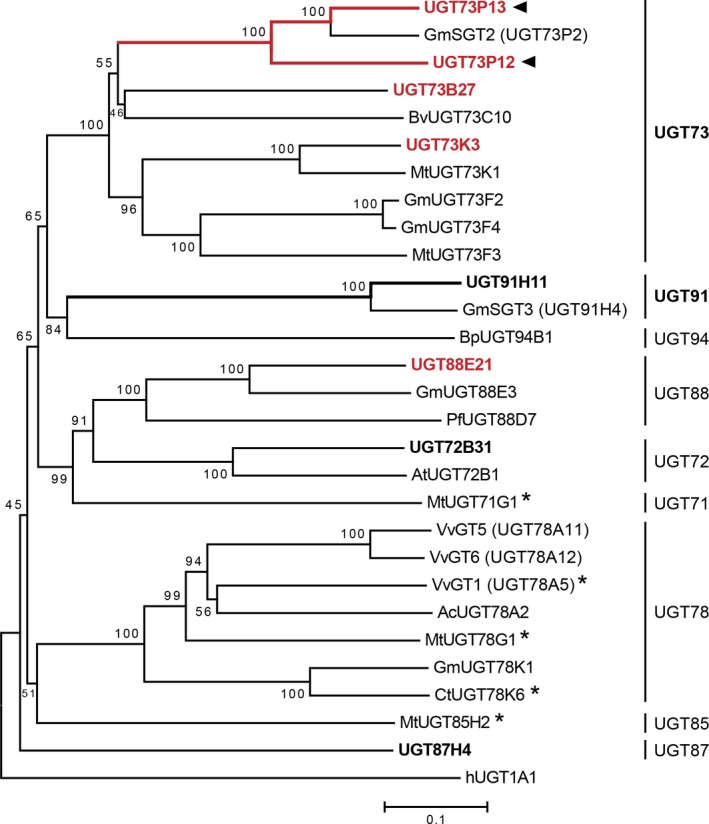
Phylogenetic relationships between candidate UGTs in Glycyrrhiza uralensis and their close relatives.
The bar indicates 0.1 amino acid substitution per site. The numbers shown next to branches are the percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates). The UGTs shown in bold font (black or red) are G. uralensis UGTs named in this study, and those shown in bold red font were further used to screen for functional proteins. Arrowheads indicate the UGTs characterized in this study. The names of close relatives that were previously characterized in other plants are tagged with the name of species abbreviated as follows: Gm, Glycine max; Bv, Barbarea vulgaris; Mt, Medicago truncatula; Bp, Bellis perennis; Pf, Perilla frutescens; At, Arabidopsis thaliana; Vv, Vitis vinifera; Ac, Aralia cordata; and Ct, Clitoria ternatea. The human (h) UGT1A1 was used as the outgroup. Superscript stars indicate the UDP‐glucose:flavonoid glucosyltransferases whose crystal structures are available.
