Figure 6.
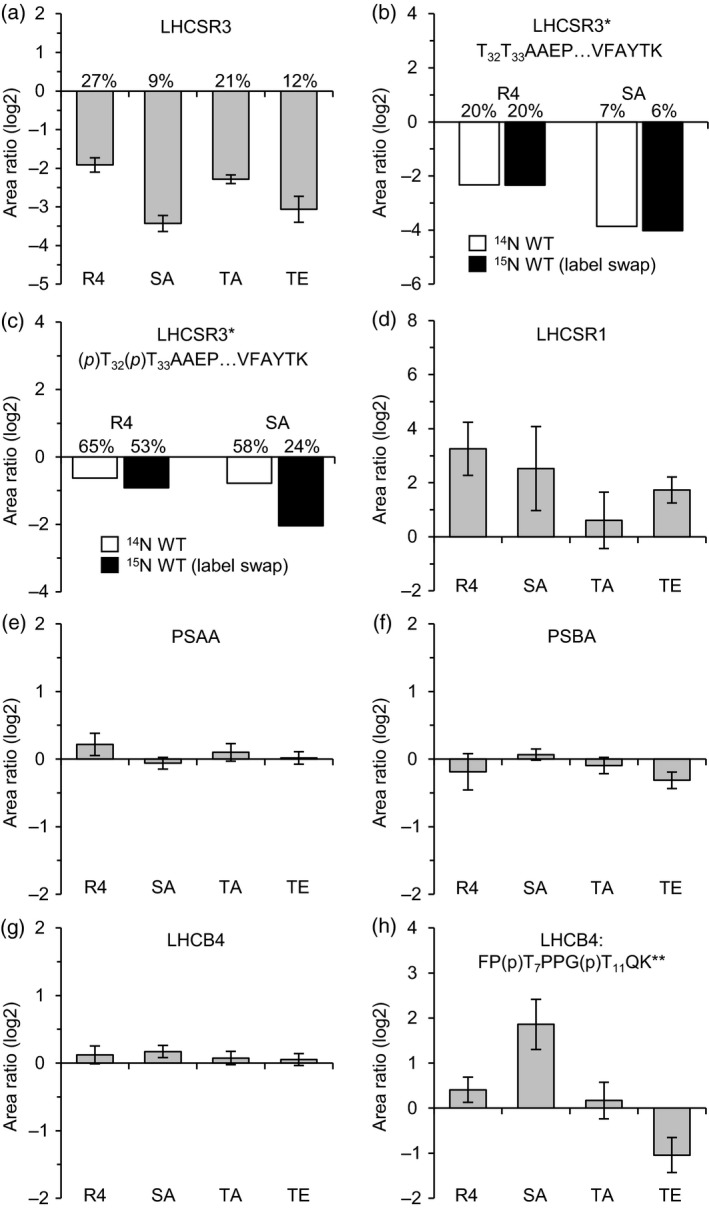
Changes in relative protein/peptide abundances and phosphorylation levels in LHCSR3 mutant and rescued strains. Unlabelled (14N) WT and labelled (15N) mutant/rescue strains were cultivated in HSM and exposed to high light (200 μmol photons m−2 sec−1) for 24 h. A replicate experiment was performed with swapped isotopic labels (15N WT, 14N mutant/rescue). Data were obtained by LC‐MS/MS (PRM) and represent the mean log2‐transformed mutant/rescue to WT peptide ratios. A minimum of three peptides was quantified per experiment. Error bars indicate the standard deviation between all peptide ratios of both replicates. Percentages denote expression levels in comparison to the wild type and were derived from mean peptide ratios prior to log2‐transformation. R4, LHCSR3‐R4; SA, LHCSR3‐S26A/S28A; TA, LHCSR3‐T32A/T33A; TE, LHCSR3‐T32E/T33E; *, quantified only in samples treated with trypsin; **, quantified only in samples treated with lysC; (p), phosphorylated residue. (a) LHCSR3. (b, c) Quantification results of the singly phosphorylated (b) and nonphosphorylated (c) versions of the LHCSR3 peptide T32T33AAEPQTAAPVAAEDVFAYTK. Data from replicate experiments are shown individually. (d) LHCSR1. (e) Photosystem I P700 chlorophyll a apoprotein A1 (PSAA). (f) Photosystem II protein D1 (PSBA). (g) LHCB4. (h) Doubly phosphorylated LHCB4 peptide FP(p)T7PPG(p)T11QK.
