Abstract
PICO (Population/problem, Intervention, Comparison, and Outcome) is widely adopted for formulating clinical questions to retrieve evidence from the literature. It plays a crucial role in Evidence-Based Medicine (EBM). This paper contributes a scalable deep learning method to extract PICO statements from RCT articles. It was trained on a small set of richly annotated PubMed abstracts using an LSTM-CRF model. By initializing our model with pretrained parameters from a large related corpus, we improved the model performance significantly with a minimal feature set. Our method has advantages in minimizing the need for laborious feature handcrafting and in avoiding the need for large shared annotated data by reusing related corpora in pretraining with a deep neural network.
Keywords: Natural Language Processing, Evidence-Based Medicine, Randomized Controlled Trial
Introduction
Evidence-Based Medicine (EBM) is the conscientious, explicit, judicious and reasonable use of modern, best evidence in making decisions about the care of individual patients [1]. However, the evidence base has been growing exponentially. It is practically impossible to catch up with the explosion of the biomedical scientific literature and realize fast and effective evidence retrieval and decision making for EBM practitioners [2; 3]. Evidence adoption at clinical practice remains suboptimal due to poorly formulated clinical questions, ineffective evidence search strategies, and disconnected databases preventing access to the best evidence.
Successfully retrieving relevant evidence begins with a well-structured question. Thus, the ability to question formulation is fundamental to locate and synthesize related resources. PICO (Population/problem, Intervention, Comparison, and Outcome) is widely adopted for formulating clinical questions to retrieve evidence from the literature. PICO stands for :
P – Population/Problem. What are the most critical characteristics of the enrolled population? What is the primary disease?
I – Intervention. What is the primary intervention considered?
C – Comparison. To what the intervention is compared?
O – Outcome. What are the anticipated measures, improvements or effects?
The PICO framework is specialized to help break down the need for evidence into searchable keywords and to formulate answerable research questions [4]. A prior study has shown that utilization of the PICO framework can improve evidence search against PubMed [5]. However, due to high demands for technical skills and medical domain knowledge for using PICO, practitioners and the general public who require searching evidence may find it either time consuming to incorporate into their busy clinical workflow, or difficult to learn. Automatic extraction of PICO statements in the biomedical literature is desired to facilitate evidence retrieval, appraisal and synthesis by clinicians and the public [6; 7].
Natural language processing (NLP) in particular promises to help us achieve this goal. Previous work has explored the use of NLP techniques to identify PICO elements in biomedical text. During the last decade, the primary solutions have evolved from knowledge-based to statistical-based such as Support Vector Machine (SVM) and Conditional Random Field (CRF) [8–11]. However, this area has attracted less attention than it should have from the NLP community, primarily caused by the lack of publicly available, annotated corpora [12], and systems almost all heavily rely on laborious handcrafted features including those specifically designed to incorporate domain knowledge.
In practice, there also lacks modularized fundamental NLP tools to support different aspect of evidence synthesis and EBM, such as tools for Named Entity Recognition (NER) to recognize PICO elements and their attributes in literature for indexing, information extraction (IE) systems for parsing and structuring study design and results from free-text literature, as well as information retrieval (IR) tools based on the PICO framework to support effective searching in literature.
With rapid advances in neural network and deep learning, recent state-of-the-art NLP systems have been developed using neural models, including some for the biomedical domain. For the Named Entity Recognition (NER) task, the best performance is achieved by biLSTM-CRF [13–15]. And transfer learning attracts increasing attention to solve high demand of large data for training neural networks [16; 17]. Recently a corpus of 5000 RCT abstracts with multi-level annotations of Patient, Intervention, and Outcomes was published, enabling new NLP application development for EBM research [12].
Compared to prior work, our PICO extraction method makes the following three significant and innovative contributions. First, it is the initial publicly available open-source NLP system for recognizing PICO elements and their attributes/measures in RCT abstracts. PICO elements are normalized with UMLS CUIs (https://github.com/Tian312/PICO_Parser). Second, this tool is developed with the minimum human labor but achieves comparable and even better performance in some categories: only a small size of gold standards is created with high inter-annotator agreement; only word feature, and no laborious handcrafted features, is used. Third, we contribute a method to reuse a large related corpus [12] under annotation guidelines different from ours to improve our model performance.
Methods
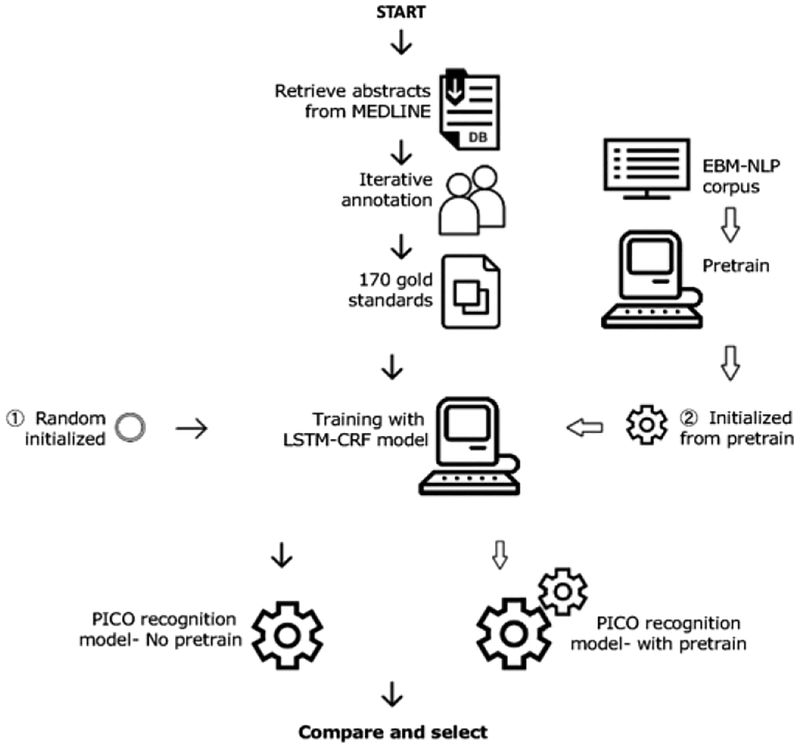
Our PICO statement extraction tool processes RCT literature following these steps: 1) Named Entity Recognition for PICO elements and attributes; 2) UMLS encoding; 3) XML output formatting. An overview of our workflow to develop the model and tool is shown in Figure 1.
Figure 1 -.
Overview of the PICO recognition tool development. We compared two optional ways for training the LSTM-CRF model (in blue): 1) with random initialization of model parameters (green, on the left); 2) “pretrain” the model with the same architecture on EBM-NLP corpus, resulting in a better parameter initialization.
Data Collection
Small Size of Gold Standards from Manual Annotation
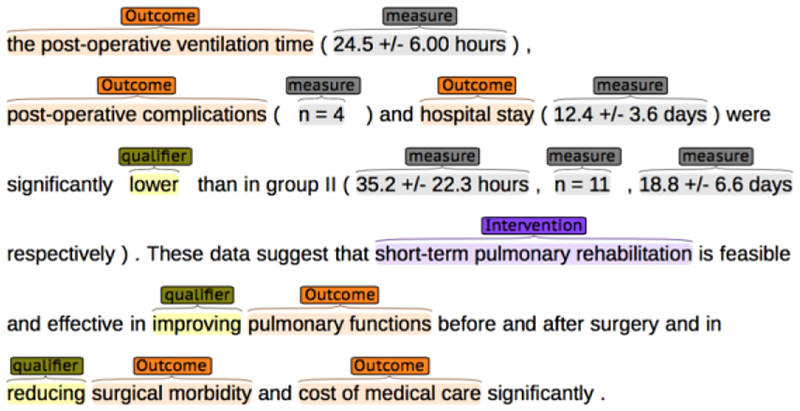
We randomly retrieved 170 RCT publications using indexed metadata from the MEDLINE database. Abstracts were retrieved from the articles and prepared in brat, a web server based collaborative annotation tool [18]. One medical professional (ZS) and one informatic researcher (TK) designed the annotation guideline for entity and attribution using an iterative process. Entity classes included in the annotation: Population, Intervention (Comparison is merged with Intervention as a subclass), and Outcome, each strictly following standard definition from the PICO framework [4]. The context for PICO elements consists of 2 types of attributes: Qualifier, a qualitative description of PICO elements (e.g., “difference”, “similar”, “higher”), and Measure, a quantitative description of PICO elements (e.g., “138 +/− 13 mg daily”). During the annotation process, both annotators followed the guideline for asking answerable research question [19]: each RCT abstract is first classified into one of 5 common clinical question types: Treatment, Prevention. Diagnosis, Prognosis, and Etiology, then annotated with PICO elements based on research type. Attributes are also identified in order to form PICO statements in entity-operator-value triplets. Each abstract is annotated at least twice by two annotators in order to ensure it strictly follows the guideline. As a rule, annotators skip annotating background and implication sections in abstracts since those parts do not usually describe study design or report objective results. An example annotation interface in brat is shown in Figure 2. This step is aimed to create a small size of annotation with a high inter-annotator agreement and high quality, serving as gold standards and core training set.
Figure 2 –
Example of our annotation in brat
A Related, Large Publicly Available Corpus
A corpus of 5000 abstracts with multi-level annotation of PIO (C is categorized as a subclass in I as well) has recently been published by a group of EBM researchers [12] (referred as EBM-NLP corpus later in this paper). The annotation was generated primarily by laypersons from Amazon Mechanical Turk (AMT) and a small part by medical professionals. The average inter-annotator agreement is measured by F1 score as 0.3, 0.18, 0.1 in span annotation and 0.5, 0.6, 0.69 in the hierarchical annotation for P, I, O classes, respectively. After reviewing this corpus, we decided that this annotated corpus cannot be directly used for our task for two major reasons. First, the annotation guidelines, primarily in part of defining element boundary and granularity, are different. In the EBM-NLP corpus, identified PICO tend to be the longest description within a sentence. While our guideline is designed to break down abstracts into the most basic elements, which can be used as “building block” for PICO statements and encoded to represent study design and results of each RCT article. For example, “Seventy-two consecutive anti-HBe-positive chronic hepatitis B patients (59 male and 13 female, median age 41 yr)” (PMID 10235220) was annotated as one Population element in EBM-NLP. While in our annotation, we recognize “consecutive anti-HBe-positive chronic hepatitis B”, “male”, “female” and “median age” as 4 independent Population entities, and “59”, “13”, “41yr” as measures. Second, as aforementioned, the EBM-NLP corpus combines measured values descriptive statistics with the PICO terms, while we have separated classes. Instead of directly training on this corpus, we believe it can be helpful for modeling a similar context as a pretrain and guide the next model training on our small gold standards.
Base model learning and pretraining
We model the task to identify PICO statements, which comprise PICO elements and their attributes in the biomedical literature, as a sequence labeling task for Named Entity Recognition (NER). NER is fundamental in general text mining, as well as in biomedical domain, e.g., recognizing problems, drug names in clinical notes, or protein, gene names in literature. As deep learning based approaches to this tasks have been gaining attention in recent years, NLP researchers now tend to prefer those methods over traditional models alone such as Support Vector Machine (SVM) or Condition Random Field (CRF) since the parameters can be learned end-to-end without the need for hand-engineered features [15]. This is particularly true in biomedical domain where traditional biomedical NLP systems heavily rely on hand-made rules and ontologies in order to reach a good performance. Deep learning methods also start attracting biomedical NLP researchers. However, these approaches are usually built upon large, high-quality labeled data, which is expensive to obtain especially in the biomedical domain because labeling biomedical text requires special medical training.
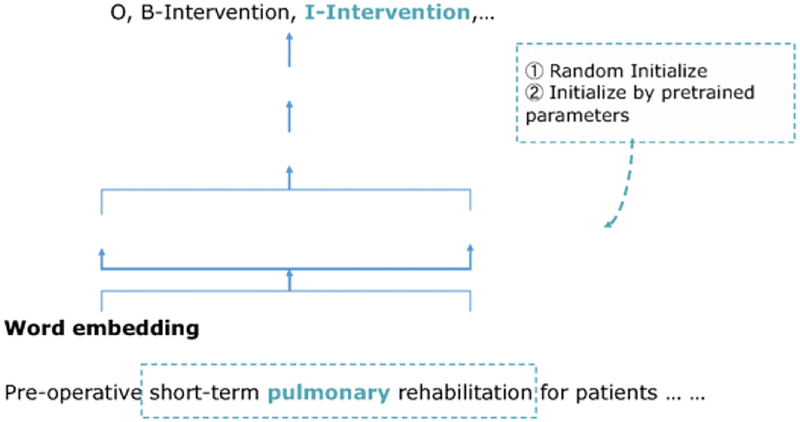
To address the lack of training corpus, recent researches start focusing on training multi-task models [20], and conducting data augmentation or transfer learning [16; 17]. Inspired by their work, we explore the feasibility and the potential way to overcome such two challenges (i.e., hand-engineered features and large, high-quality data) in a small training set and simple feature with the help of the public data. We adopt the bidirectional Long short-term memory (LSTM), a kind of recurrent neural network as our base model, and decode with a linear chain CRF in the output layer (biLSTM-CRF). This architecture now achieves state-of-the-art performance in NER tasks. The model details are illustrated in Figure 3. We use classical “BIO” tags to represent the boundary of terms of interest: “O” means it is outside the target terms. “B” represents the beginning word, and “I” tags all the inside words. We compare the results trained with raw tagging to BIO tagging methods. Tagged output for the example in Figure 3 is:
Figure 3 -.
Base model detailed architecture. It’s used to train both PICO recognition model and EBM-NLP corpus.
Pre-operative/B-Intervention short-term/I-Intervention pulmonary/I-Intervention rehabilitation/I-Intervention for patients …
The base model is similar to [14], and is also used in EBM-NLP corpus paper to generate task baseline for identifying PICO span. The dark green in Fig. 3 represents modules learned during training. While the light green, namely word embedding is pretrained on an entire collection of abstracts on PubMed from 1990–2018 using word2vec toolkit [21]. During the learning phase, the model first generates a character-based representation of each word and concatenate it with pretrained word vectors. In other words, each input word is represented by two concatenated vectors in both word level and character level. In the next step, each sentence as sequenced word vectors is then fed into a bidirectional LSTM to extract the contextual representation of each word.
At this step, we can get a likelihood at word level through a decoder layer. A significant drawback of optimizing by word-level likelihood is that it doesn’t consider dependencies between neighboring in the sentence. Thus, a CRF layer is added to model the entire sentence structure. CRF is a log-linear graphical model that additionally considers the transition score from one tag to the next. This characteristic makes it a classic model in traditional NER tasks. After decoding by CRF, the log likelihood is maximized for the entire sentence in order to select the best tag for the target word. Like in Figure 3, when the target word is “pulmonary”, all the neighbor words in a window are considered to generate the tag. Only word features, i.e., word vectors and character vectors, are used in this model, without any feature engineering.
Without pretraining, biLSTM in the base model is randomly initialized and then optimized using the Adam optimizer. For pretraining, we use the entire 5000 abstracts in EBM-NLP corpus with “starting span” annotation (only in PICO level, no further hierarchical labeling) in the same model architecture, and then transfer the learned weights to the PICO recognition model. Next, we fine-tune the PICO recognition model to reach the best performance. All models are trained using TensorFlow (https://www.tensorflow.org/).
Concept Normalization and Output Structuring
We select the best model as the backend support of our PICO recognition tool. Given one or a set of free text abstracts as input, the tool automatically recognizes Patient, Intervention (including Comparison), Outcome elements and corresponding attributes. In order to support further computational tasks, the recognized PICO elements are encoded with the Unified Medical Language System (UMLS, https://www.nlm.nih.gov/research/umls/), an integrated biomedical terminology, by applying a UMLS concept extraction tool QuickUMLS [22]. Extracted semantics are further organized into a structured format. The default output format is XML, while users can also choose JSON, as more recently published APIs use JSON as standard data format.
Results
Descriptive Statistics of the Annotated Corpus
We created a sharable, finely annotated corpus for PICO extraction with its descriptive statistics provided in Table 1.
Table 1.
Descriptive statistics of the annotated corpora
| Entity class | Attribute class | ||||
|---|---|---|---|---|---|
| P. | I. (+C.) | O. | Qualifier | Measure | |
| Count | 1185 | 2027 | 2140 | 766 | 904 |
| Agreement | 0.916 | 0.844 | 0.727 | 0.955 | 0.954 |
The inter-annotator agreement is evaluated by cohen’s κ statistic. The overall agreement between the two annotators for 5 categories is 0.83. The category-specific κ measures is reported in Table 1. Our goal in this step is to create a corpus of high-quality annotation with high agreement. Thus, our annotation team spent much time on iterative annotation guideline design and test run in sample corpus for multiple rounds to resolve discrepancies between annotators and arrive at consensus understandings for each class and required granularity. Compared to the related NLP work, we have a relatively small corpus to minimize human labors. We plan to achieve satisfactory performance with such small corpus.
Model Performance
For evaluation purpose, 6-fold cross-validation is applied. 170 abstracts are equally divided into 6 groups. Among each run of model learning and testing, 4/6 of the data used as training set, 1/6 as validation set and 1/6 as test set. We report the performance on test sets.
Classic evaluation metrics are generated for evaluating NER tasks: precision, recall and, F1 score, to evaluate model performance in two different levels: word level and token level and use represent the two by span and trunk. In word level or span evaluation, the basic unit is the word, while in trunk evaluation, basic unit is a token. Using an intervention element with BIO tagging as an example, “short-term/B-Intervention pulmonary/I-Intervention rehabilitation/I-Intervention”, in span evaluation, there are 3 predictions for each word. A true positive is counted when both BI tag and class are predicted correctly. There are 3 true positives at most. While in trunk evaluation, “short-term pulmonary rehabilitation” is counted as one token, 1 true positive is counted only if both boundary and class of this token are correctly predicted.
The model performance is reported based on the test set in Table 2. We test performance with different tagging methods (raw/BIO tagging) and pretrain or not. For each model setting, we report the best evaluation among 6 sets for cross-validation and also averaged measures. In summary, using BIO tagging and pretrain can both improve model performance. In word-level, span evaluation, the best performance comes from the model using pretrain and raw tagging with 0.78 in averaged F1 score, and the best F1 in the subset is 0.89. Compared to the model setting with raw tagging as well but using no pretrain, the F1 score has been improved about 10%. With pretrain and BIO tagging, the performance is also improved, but not as much as using raw tagging. It’s a reasonable result as in BIO tagging a word is counted as true positive requiring both BI tags and class are correct while raw tagging only require class prediction. On the other hand, BIO tagging provides more We further analyzed the individual performance from one of the 6 sets using the best model setting (Pre+BIO). The details are shown in Table 3. As evaluated by F1 score, entities with B tags are generally better than I tags, indicating the model is better at predicting if there is an entity, but need to be improved to predict the span of it. I tag prediction is especially poor in evaluation for Modifier, with F1 score only 0.25. We retrieved raw prediction results for Modifier class. We found due to the fact that we have a small gold standard set, and only 1/6 used for testing, there are only 166 Modifier tokens in the test set, among which only 6 are not unigram (have I tags). Modifiers are usually one-word token such as “higher”, “rise”, and “similar”. Among 6 modifiers with I tags in the test set, the model predicts 2 I-modifier tags and 1 of them is correct. Thus, precision/recall is 0.5/0.16 and F1 is calculated as low as 0.25, but actually caused by its small total number in entire corpus.
Table 2.
Model performance in different training settings.
| No Pre + Raw | No Pre + BIO | Pre + Raw | Pre + BIO | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Best | Ave. | Best | Ave. | Best | Ave. | Best | Ave. | ||
| Test set (Span) | Precision | 0.78 | 0.76 | 0.84 | 0.85 | 0.93 | 0.87 | 0.86 | 0.83 |
| Recall | 0.62 | 0.63 | 0.68 | 0.66 | 0.80 | 0.70 | 0.71 | 0.7 | |
| F1 score | 0.69 | 0.66 | 0.75 | 0.74 | 0.89 | 0.78 | 0.78 | 0.73 | |
| Test set (Trunk) | Precision | 0.54 | 0.52 | 0.58 | 0.53 | 0.74 | 0.61 | 0.63 | 0.63 |
| Recall | 0.53 | 0.51 | 0.56 | 0.52 | 0.74 | 0.56 | 0.64 | 0.61 | |
| F1 score | 0.53 | 0.52 | 0.57 | 0.54 | 0.74 | 0.58 | 0.64 | 0.62 | |
Table 3.
Detailed evaluation for one set in PICO/attribute
| B-Pop. | I-Pop. | B-Int. | I-Int. | B-Out. | I-Out. | |
|---|---|---|---|---|---|---|
| Precision | 0.82 | 0.84 | 0.82 | 0.78 | 0.88 | 0.85 |
| Recall | 0.68 | 0.65 | 0.70 | 0.50 | 0.75 | 0.42 |
| F1 score | 0.75 | 0.74 | 0.75 | 0.61 | 0.81 | 0.56 |
| B-Mea. | I-Mea. | B-Qua. | I-Qua. | |||
| Precision | 0.77 | 0.85 | 0.91 | 0.5 | ||
| Recall | 0.65 | 0.65 | 0.60 | 0.17 | ||
| F1 score | 0.71 | 0.74 | 0.72 | 0.25 |
Sample output
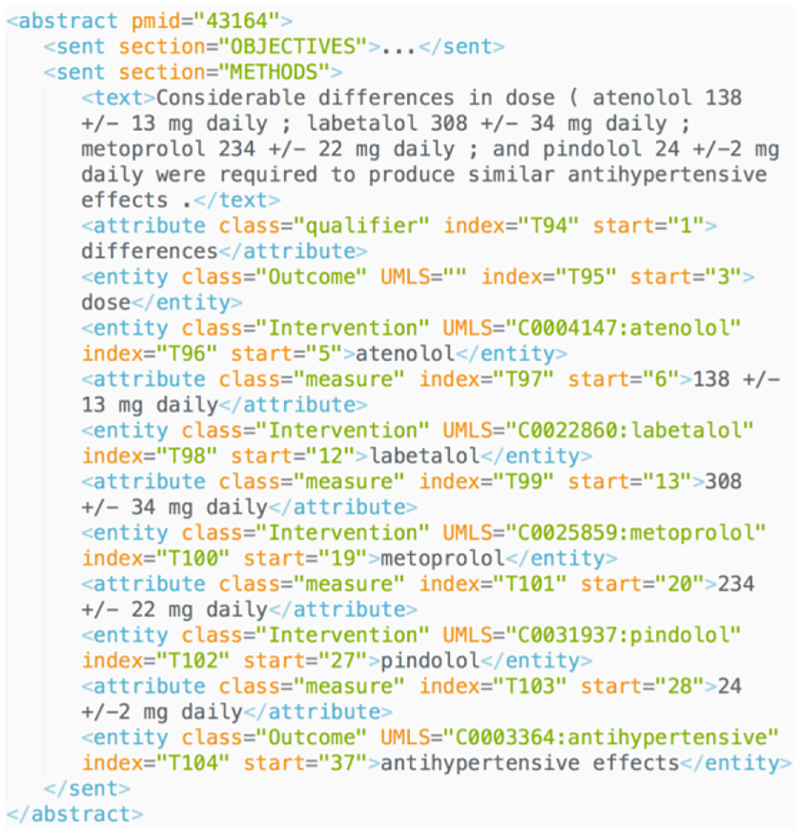
The models are trained with following parameters: mini batch (size of 5) and Adam optimizer are selected for training; the dimensions of the word and character vectors are 200 and 100; the learning rate is set as 0.001with a decay of 0.9. Pretrain converges within 50 epochs and the training on 170 abstracts within 10 epochs. A sample recognition result in XML format is shown in Figure 4 It contains rich parsed semantic and positional information that can support further computational tasks such as relation extraction and information retrieval. Sample JSON output can be found in our github repository. information for learning to help identify the boundary of each element. This is reflected by the evaluation in trunks/token level. The best performance in token level is generated by model with pretrain and BIO tagging (average F1 score 0.62, best 0.64). Compared to the two model only trained on 170 abstracts (0.52/0.54 for average F1), pretraining on EBM-NLP corpus and transferring learned parameters also help improve the model performance significantly by 10%. Therefore, applying pretrain and BIO tagging can best improve the recognition of PICO elements boundaries and predicting PICO classes.
Figure 4 -.
Sample output for our PICO extraction method
Discussion
Error Analysis
The most common error happens when multiple PICO terms appear in conjunction. For example, an RCT paper titled “Perioperative enteral nutrition and quality of life of severely malnourished head and neck cancer patients: a randomized clinical trial”, and one Population entity recognition result is:
<entity class=Population UMLS=‘C0278996:head and neck cancer,C0162429:malnourished,C0205082:severely’ index=‘T3’ start=‘8’> severely malnourished head and neck cancer </entity>
Although we define the PICO elements to be the annotated as the most basic concepts (should be “malnourished” and “head and neck cancer” the two P entities in this case), there was variance in what annotators considered, also causing inconsistency when calculating inter-annotator agreement.
Comparative Performance Evaluation Results
In the EBM-NLP corpus, the best performance for the baseline model trained on 5000 abstracts for P, I, O classes are 0.71, 0.65 and 0.63 by F1 score respectively (mathematical mean: 0.66) at the word level. In contrast, with a small gold standard set (170 abstracts) and without any hand-engineered features, our model reaches 0.78 for the best average F1 score at word level and 0.62 at the token level. Our results prove the effectiveness of pretraining in minimizing human efforts in annotation and features engineering while reaching satisfactory performance.
Future Work
We have not yet related the attributes to their PICO elements nor distinguished PICO elements by arms. To further complete the structured information, negation and semantic relations need to be identified. We will progressively complete the functions of this tool, and eventually turn it to comprehensive information extraction system to computationally represent abstracts describing RCTs.
Conclusions
In this study, we demonstrate the early promise of pretraining to improve model performance tuned on a small training set, with only word feature, and we achieve better performance than conventional machine learning models trained on a larger corpus. This result is significant in showing the feasibility of overcoming the challenges in the dearth of annotated data and laborious feature handcrafts in biomedical NLP. We also contribute an open source NLP tool to automatically recognize PICO elements and their attributes from RCT abstracts. This tool, can be used to structure study design and results and can further enhance evidence retrieval and synthesis from biomedical literature to facilitate evidence-based medicine.
Acknowledgments
This project was supported by USA NIH grant R01LM009886-08A1 (Bridging the semantic gap between research eligibility criteria and clinical data; PI: Weng).
References
- [1].Masic I, Miokovic M, and Muhamedagic B, Evidence based medicine - new approaches and challenges, Acta Inform Med 16 (2008), 219–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bastian H, Glasziou P, and Chalmers I, Seventy-five trials and eleven systematic reviews a day: how will we ever keep up?, PLoS medicine 7 (2010), e1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Fraser AG and Dunstan FD, On the impossibility of being expert, Bmj 341 (2010), c6815. [DOI] [PubMed] [Google Scholar]
- [4].Richardson WS, Wilson MC, Nishikawa J, and Hayward RS, The well-built clinical question: a key to evidence-based decisions, ACP journal club 123 (1995), A12–A12. [PubMed] [Google Scholar]
- [5].Schardt C, Adams MB, Owens T, Keitz S, and Fontelo P, Utilization of the PICO framework to improve searching PubMed for clinical questions, BMC medical informatics and decision making 7 (2007), 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Fontelo P, Liu F, and Ackerman M, ask MEDLINE: a free-text, natural language query tool for MEDLINE/PubMed, BMC medical informatics and decision making 5 (2005), 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wallace BC, Dahabreh IJ, Schmid CH, Lau J, and Trikalinos TA, Modernizing the systematic review process to inform comparative effectiveness: tools and methods, Journal of comparative effectiveness research 2 (2013), 273–282. [DOI] [PubMed] [Google Scholar]
- [8].Demner-Fushman D and Lin J, Answering clinical questions with knowledge-based and statistical techniques, Computational Linguistics 33 (2007), 63–103. [Google Scholar]
- [9].Kim SN, Martinez D, Cavedon L, and Yencken L, Automatic classification of sentences to support evidence based medicine, in: BMC bioinformatics, BioMed Central, 2011, p. S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Marshall IJ, Kuiper J, Banner E, and Wallace BC, Automating biomedical evidence synthesis: RobotReviewer, in: Proceedings of the conference. Association for Computational Linguistics. Meeting, NIH Public Access, 2017, p. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wallace BC, Kuiper J, Sharma A, Zhu M, and Marshall IJ, Extracting PICO sentences from clinical trial reports using supervised distant supervision, The Journal of Machine Learning Research 17 (2016), 4572–4596. [PMC free article] [PubMed] [Google Scholar]
- [12].Nye B, Li JJ, Patel R, Yang Y, Marshall IJ, Nenkova A, and Wallace BC, A Corpus with Multi-Level Annotations of Patients, Interventions and Outcomes to Support Language Processing for Medical Literature, arXiv preprint arXiv:1806.04185 (2018). [PMC free article] [PubMed] [Google Scholar]
- [13].Lample G, Ballesteros M, Subramanian S, Kawakami K, and Dyer C, Neural architectures for named entity recognition, arXiv preprint arXiv:1603.01360 (2016). [Google Scholar]
- [14].Ma X and Hovy E, End-to-end sequence labeling via bi-directional lstm-cnns-crf, arXiv preprint arXiv:1603.01354 (2016). [Google Scholar]
- [15].Sachan DS, Xie P, Sachan M, and Xing EP, Effective Use of Bidirectional Language Modeling for Transfer Learning in Biomedical Named Entity Recognition.
- [16].Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, and Kang J, BioBERT: pre-trained biomedical language representation model for biomedical text mining, arXiv preprint arXiv:1901.08746 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Giorgi JM and Bader GD, Transfer learning for biomedical named entity recognition with neural networks, Bioinformatics 34 (2018), 4087–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Stenetorp P, Pyysalo S, Topić G, Ohta T, Ananiadou S, and Tsujii J.i., BRAT: a web-based tool for NLP-assisted text annotation, in: Proceedings of the Demonstrations at the 13th Conference of the European Chapter of the Association for Computational Linguistics, Association for Computational Linguistics, 2012, pp. 102–107. [Google Scholar]
- [19].Fineout-Overholt E and Johnston L, Teaching EBP: Asking searchable, answerable clinical questions, Worldviews on Evidence-Based Nursing 2 (2005), 157–160. [DOI] [PubMed] [Google Scholar]
- [20].Wang X, Zhang Y, Ren X, Zhang Y, Zitnik M, Shang J, Langlotz C, and Han J, Cross-type biomedical named entity recognition with deep multi-task learning, arXiv preprint arXiv:1801.09851 (2018). [DOI] [PubMed] [Google Scholar]
- [21].Mikolov T, Chen K, Corrado G, and Dean J, Efficient estimation of word representations in vector space, arXiv preprint arXiv:1301.3781 (2013). [Google Scholar]
- [22].Soldaini L and Goharian N, Quickumls: a fast, unsupervised approach for medical concept extraction, in: MedIR workshop, sigir, 2016. [Google Scholar]