Abstract
Section Title
Breast cancer is the most common cause of death among women worldwide. Although there are many known risk factors in breast cancer development, infectious diseases have appeared as one of the important key to contribute to carcinogenesis formation. The effects of Human Cytomegalovirus (HCMV) on women with breast cancer has been recently studied and reported. To contribute to this research trend, this study was conducted to evaluate the association between HCMV and the women with breast cancer.
Objective:
This experiment aimed to evaluate HCMV DNA in women with breast cancer in Ahvaz city, Iran.
Materials and Methods:
A total of 37 formalin fixed paraffin embedded tissues of the patients with ductal breast carcinoma and 35 paraffin embedded tissues of the patients with fibro adenoma as control group were collected. The deparaffinization of all the samples were carried out and the DNA was extracted. Initially, the PCR test was carried out to detect beta –globulin DNA as an internal control. For those samples positive for beta –globulin DNA, Polymerase Chain reaction (PCR) was used to detect HCMV for the tests and control samples.
Results:
Among 37 ductal breast carcinoma, 20 (54.04%) cases were proved positive for HCMV DNA by PCR. While among the 35 control group (fibroadenoma), 10 (28.57%) cases were positive for HCMV DNA (P >0.028). The prevalences of HCMV DNA among the age groups 30-39, 40-49 and >50 years were 7 (72.22%), 9 (69.23%), 4 (57.14%), respectively (P=0.066). A high frequency of HCMV DNA was detected in tumor grade III, 13/18 (58.33%) compared with tumor grade II, 7/19 (36.84%) (p=0.044). A high frequency of 16/24 (66.66%) of HCMV DNA was found in invasive ductal breast cancer compared with 4/13 (30.76%) HCMV DNA in situ (P<0.028).
Conclusion:
A high prevalence of 54.05% HCMV was found among the patients with ductal carcinoma. The percentages of the high prevalence of HCMV among age group (40-49) years, tumors grades, and invasive stage were (69.23%), (58.33%), (66.66%), respectively. Further study of HCMV in the latency phase in patients with ductal carcinoma would be necessary to extend our knowledge.
Key Words: Human cytomegalovirus, breast cancer, ductal carcinoma, Polymerase Chain Reaction
Introduction
Breast cancer is a leading cause of death among women worldwide. There are several risk factors for breast carcinoma such as early age of menarche, late age of menopause, a positive family history of breast cancer, hormone replacement therapy, age, sex, and nulliparity (Ban et al., 2014; Jemal et al., 2011; Anders et al., 2009). The prevalence of breast cancer has been frequently reported in Iran (Sajad et al., 2009; Mohammadizadeh et al., 2014; Karimi et al., 2016; Mohammadizadeh et al., 2017). So far, investigations have implied a number of different viral infections associated with breast cancer, including bovine leukemia virus (Buehring et al., 2015). Human mammary tumor virus (Melana et al., 2010), human papillomavirus (Heng et al., 2009), Epstein–Barr virus (EBV) ( Joshi et al., 2009; Fawzy et al., 2008; Hachana et al., 2011), polymavirus (Antonsson et al., 2012) and human cytomegalovirus (HCMV) (Karimi et al., 2016; El-Shinawi et al., 2013; Richardson et al., 2015; Mohammadizadeh et al., 2017). Although the role of these viruses in the breast cancer are arguable, the evidence of molecular epidemiological suggests an association between HCMV and breast cancer. HCMV is a β-herpesvirus that affects 70–90% of the world population, causing general, acute, persistent, or lifelong latent infection (Goodrum et al., 2012). HCMV infections are typically subclinical and serious diseases which occur chiefly in immune-compromised individuals (Dziurzynski et al., 2011). Overall, HCMV serostatus has not been positively associated with breast cancer. However, women with breast cancer were found to have higher mean HCMV IgG levels in an Australian case–control study suggesting that they might have afflicted a recent infection (Richardson et al., 2004). Although HCMV proteins and DNA have been detected in breast tumor tissue, HCMV is not typically considered as an oncogenic virus (Michaelis et al., 2009). HCMV can promote many classic signs of cancer, such as cell cycle dysregulation, inhibition of apoptosis, increased migration and invasion, and immune evasion (Sanchez et al., 2008). HCMV has been associated with other malignancies, including glioblastoma (Soroceanu et al., 2011), medulloblastoma (Baryawno et al., 2011), colon cancer (Tafvizi et al., 2014), prostate cancer (Samanta et al., 2003) and Hodgkin and Non-Hodgkin Lymphoma (Hamide et al., 2017). Individual HCMV gene products can have profound effects on cell growth, such as immediate early proteins (IE1) and IE2, which are known to stimulate entry into S phase (Castillo et al., 2002). IE1 expression was observed to increase the growth rate of glioblastoma cells in culture, suppress p53 and Rb tumor suppressor activity, and stimulate PI3K/Akt signaling (Cobbs et al., 2008). IE1 was detected in breast tumor tissue (Harkins et al., 2010; Taher et al., 2013). Another HCMV gene, US28 displays constitutive signaling activity, and cells expressing US28 are highly invasive and form tumors in nude mice (Soroceanu et al., 2011; Maussang et al., 2009). US28 was shown to induce vascular endothelial growth factor (VEGF), cyclooxygenase-2 (COX2), and STAT3 activation through up-regulation of IL-6 (Maussang et al., 2009; Slinger et al., 2010). The HCMV UL111A gene encodes cmvIL-10, a viral cytokine which is secreted from infected cells (Jones et al., 2002). Extensive immunosuppressive properties of cmvIL-10 was found to down regulate class I and II MHC, and inhibit the dendritic cells maturation (Slobedman et al., 2009; Chang et al., 2009; Avdic 2014). Engagement of the IL-10 receptor by cmvIL-10 leads to activation of STAT3 which is commonly activated in breast cancer cells (Banerjee et al., 2016). It was found that STAT3 activation is associated with poor prognosis in ovarian cancer and is considered as a key factor in metastasis formation (Zhang et al.,2010). With the aforementioned properties of HCMV and its association with different human cancers, this study was conducted to evaluate the HCMV DNA in the formalin-fixed paraffin-embedded tissues of the patients with ductal breast carcinoma in Ahvaz city. Ahvaz city is capital of Khuzestan province with the population of 1.5 million people, located in the southwest of Iran.
Materials and Methods
Fifty blocks of the formalin-fixed paraffin-embedded tissue blocks of ductal breast cancer and forty three fibroadenoma as a control group were collected from the archive of Imam Khomeini Hospital, Ahvaz, Iran during 2006-2014. The diagnostic accuracy of ductal breast carcinoma were approved by a pathologist. The patients ages were between 40 and 59 years with the mean age of 55±8 years. The sections of 10 μm thickness were prepared from each sample and stored at 4°C until tests performance .
1-Deparaffinization: Deparaffinization was done by xylene and ethanol (Germany, Merk). Initially, all the specimens were placed in microtubes then xylene was added and kept at 45C° for 15 min followed by centrifuge at 14,000 rpm for 1 minute. This stage was repeated again. The supernatant was discarded and 1ml absolute ethanol was added to precipitate. It was stored at the room temperature for 10 min and centrifuged again at 14,000 rpm for 1 minute. The supernatant was discarded. This process was repeated by adding 70% ethanol, followed by the same condition. Finally, supernatant was discarded and all microtubes were placed at 65oC for 5 min to vaporize the ethanol residue and the pellet was used in DNA extraction (Habibian et al., 2013 ).
2-DNA extraction: High pure PCR template preparation kit (Roche, Germany, code No: 11796828001) was applied for the extraction of DNA, according to the manufacturer’s instruction. The extracted DNA was stored at -70°C until PCR amplification.
3-PCR amplification: All the extracted DNA samples were initially subjected to PCR with consensus primers PCO3/PCO4 (β-globin) to confirm the quality of the extracted DNA (used as an internal control). The following primers (PCO3: 5 ´ACACAACTGTGTTCACTAGC/PCO4: 5 ´CAACTTCATCCACGTTCACC with PCR product of 110 bp (Shahab et al., 2015). The following primers conserved for the GB region of HCMV included, F primer 5́- TCTGGGAAGCCTCGGAACG -3 (1,043-1,062) and R primer 5- GAAACGCGCGGCAATCGG-(1,621-1,604). was used to detect HCMV. The first round of PCR was performed in 25μl mixture, containing 10μl of extracted DNA, 2.5μl PCR buffer 10X (Roche),0.5μl dNTP 10mM (Roche), 1U Taq Polymerase (Roche), 1µl(20μM) of each primer sequence, D/W up to 25μl was subjected to thermocycler (Techne TC-5000, UK) (Gilbert et al.,1999). The products of 581bp indicating positive reaction.
4-Gel electrophoresis: The second round of PCR product was separated on a 2% agarose gel and developed by Safe Stain under voltage at 100V. The result was seen under ultra violet in transilluminator. The sizes of bands were compared with 100bp Ladder (Fermentas) which was placed on the well as an indicator.
Figure 1.
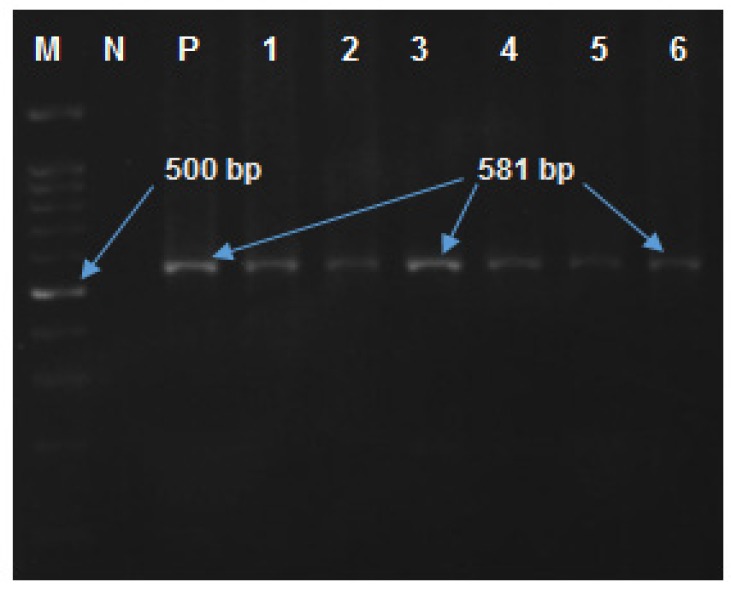
Lane M, molecular size maker, 100 bp DNA ladder; lane N, negative controls; Lane P, positive control; Lane 1-6, amplified products (581bp) on agarose gel electrophoresis
To confirm the results of PCR and to determine genotyping randomly, 5 positive PCR products were selected and sequenced (Bioneer company, South Korea). The sequences were blasted using available databases.
Statistical analysis
The obtained results were analyzed by the version 17 of SPSS software and the role of age and sex on positive cases were surveyed by Fisher`s exact and Chi square test.
Results
Among 37 ductal breast carcinoma, 20 (54.04%) cases were positive for HCMV DNA by PCR. On the other hand, among the 35 control group (fibroadenoma), 10 (28.57%) cases were positive for HCMV DNA (P >0.028). The prevalence of HCMV DNA among the age groups 30-39, 40-49 and >50 years was 7(18.91%), 9 (69.23%), 4/7 (10.81%) respectively (P=0.066). High frequency of HCMV DNA have been found in tumor grade III13/18 (72.22%) compared with tumor grade II7/19 (36.84%) (p=0.044). A high rate of 16/24 (66.66%) HCMV DNA was found in invasive ductal breast cancer in comparison with the results 4/13 (30.76%) HCMV obtained in situ (P<0.028) (Table 1).
Table 1.
Profile of Patients with Breast Cancer Type Ductal Carcinoma
| Category | HCMV positive | HCMV negative | p-value |
|---|---|---|---|
| Ages | |||
| <20 | 0/5 | 5/5 | 0.066 |
| 30-39 | 7/12 (58.33%) | 5 (41.66%) | |
| 40-49 | 9/13 (69.23%) | 4 (30.76%) | |
| <50 | 4/7 (57.14%) | 3 (42.85%) | |
| Ductal carcinoma | |||
| No:37 | 20/37 (54.05%) | 17/37 (45.94%) | 0.028 |
| Adeno | |||
| Fibroma (No:35) | 10/35 (28.57%) | 25 (71.42%) | |
| Tumor grade | |||
| Grade II | 7/19 (36.84%) | 12 (63.15%) | 0.044 |
| Grade III | 13/18 (72.22%) | 5 (27.77%) | |
| Breast cancer | |||
| Invasive N=24 | 16/24 (66.66%) | 8 (33.33%) | 0.028 |
| In situ N=13 | 4/13 (30.76%) | 9 (69.23%) | |
Table 1 shows that the distribution of HCMV among the age-group was not significant (p=0.066), while it was found significant among the ductal carcinoma and fibroma (p=0.028), tumor grade II and III (p=0.044), and invasive and in situ (p=0.028).
Discussion
Although, the association between HCMV and cancer is arguable, there are documentations which reveal that progression of some tumors could be intensified by the proteins named US28, pp65, IE1, encoded by HCMV genes during the phase of latent infection (Soroceanu et al., 2011; Maussang et al., 2009; Cai et al., 2016; Lucas, 2011). PP65 is the most abundant virion protein and non-infectious viral particles that are assembled during active infection (Libard et al., 2014). Besides, the detection of IE1mRNA indicates the presence of activation or reactivation of HCMV infection (Harwardt., 2016). The IE protein is detected in breast tumor tissue (Harkins et al., 2010; Taher et al., 2013; Mohammadizadeh et al., 2017). However, the association and expression of US28, pp65 and IEI genes were not evaluated in the present study, which requires further investigations. Additionally, there are some factors involved in the latency of HCMV. These proteins are UL133-UL138, encoded by HCMV genes and involved in regulating latency, viral immune escape and cell tropism (Petrucelli et al., 2012; Montag et al., 2011; Hamide et al., 2016). The detection of HCMV UL-133,-138 genes were not carried out in the presence study.
In the presence study, a high prevalence of 54.04% HCMV DNA was found in patients with ductal breast cancer while 28.57% of control group were positive for HCMV DNA. The status of HCMV detection in the latency phase in patients with ductal breast cancer was not clear but 20 (54.04%) patients with ductal breast carcinoma appeared positive for HCMV DNA by PCR. On the other hand, among the 35 control group samples (fibroadenoma), 10 (28.57%) cases were positive for HCMV DNA (P >0.028.). In accordance to our findings, (Karimi et al., 2016) detected HCMV DNA in 26/50 (58%) samples of invasive breast carcinoma by using the nested-PCR method in Sanandaj city, Iran.
Taher et al., (2013) from Sweden demonstrated HCMV IE protein expression in 100% of 73 breast cancer samples using the IHC method and real-time PCR. Harkins et al., (2010) in the United States also evaluated the surgical biopsy specimens of 38 normal breast samples, 39 breast carcinoma samples, and paired normal breast tissue from 21 breast cancer patients, and demonstrated a higher expression of HCMV immediate early (IE), early and late (E/L) and late (L) antigens in breast cancer 31/32 (97%) compared to normal breast epithelium 17/27 (63%).
El-Shinawi et al., (2013) from Egypt also reported a significant association between HCMV and breast cancer with higher serum levels of HCMV IgG in 82% of 28 patients with inflammatory breast carcinoma compared to 65% of 49 patients with non-inflammatory breast carcinoma.
On the contrary, several studies have not spotted any relationship between HCMV and breast cancer. Utrera-Barillas et al., (2013) evaluated 27 breast cancer specimens and 20 fibroadenoma samples by quantitative PCR and reported no significant association between HCMV and breast cancer development in Mexico. Richardson et al., (2015) from New Zealand also evaluated the CMV IgG levels in plasma (by the enzyme immunoassay method) and checked HCMV DNA in 70 tumor samples using the quantitative PCR method and found no relationship between HCMV and breast cancer.
In conclusion, this research documented a high prevalence of 54.05% for HCMV DNA among the patients with ductal carcinoma. In detail, high prevalence rates of HCMV DNA among the patients’ group (40-49) years (69.23%), tumors grades (72.22%), and invasive stage (66.66%) were detected.
In conclusion, further research is needed to investigate the HCMV role in the latency phase in patients with ductal carcinoma as the role of HCMV in the latency phase and it association with ductal carcinoma could not be justified by this experiment. To do this, the expression of UL-133-138, US28, pp65 and IEI genes needs to be evaluated. To manage and treat individuals with breast cancer prior to chemotherapy, the detection of HCMV by PCR or Real time PCR should be implemented.
Acknowledgements
This study was done as a research project with 92124 registration number in Infectious and Tropical Diseases Research Center Health Research Institute Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran. We appreciate Dr. Somayeh Biparva Haghighi, assistant professor of applied linguistics at Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran, for her precise language editing of the current article.
Authors’ Contributions
Study concept and design: Manoochehr Makvandi, Alireza Samarbafzadeh; acquisition of data: Payman Sepahv; analysis and interpretation of data:
Manoochehr Makvandi and Payman Sepahvand; drafting of the manuscript: Payman Sepahvand; critical revision of the manuscript for intellectual content: Manoochehr Makvandi; statistical analysis: Ahmadi Angali Kambiz and Payman Sepahvand; administrative, technical, and material support: Payman Sepahvand, Niloofar Neisi, Abdolhassan Talaei-Zadeh, Nastarn Ranjbari, Nilofar Nisi. Azarakh Azaran, Shahram Jalilian, Mehran Varnaseri; study supervision: Manoochehr Makvandi and Alireza Samarbafzadeh.
Funding/Support
This study was supported by Health Research Institute, Infectious and Tropical disease Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran.
Financial Disclosures
The authors have no financial interest related to the material in the manuscript.
References
- Anders CK, Carey LA. Biology, metastatic patterns, and treatment of patients with triple-negative breast cancer. Clin Breast Cancer. 2009;9:73–81. doi: 10.3816/CBC.2009.s.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonsson A, Bialasiewicz S, Rockett RJ, et al. Exploring the prevalence of ten polyomaviruses and two herpes viruses in breast cancer. PLoS One. 2012;7:e39842. doi: 10.1371/journal.pone.0039842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avdic S, McSharry BP, Slobedman B. Modulation of dendritic cell functions by viral IL-10 encoded by human cytomegalovirus. Front Microbiol. 2014;5:337. doi: 10.3389/fmicb.2014.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee K, Resat H. Constitutive activation of STAT3 in breast cancer cells:a review. Int J Cancer. 2016;138:2570–8. doi: 10.1002/ijc.29923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban KA, Godellas CV. Epidemiology of breast cancer. Surg Oncol Clin N Am. 2014;23:409–22. doi: 10.1016/j.soc.2014.03.011. [DOI] [PubMed] [Google Scholar]
- Baryawno N, Rahbar A, Wolmer-Solberg N, et al. Detection of human cytomegalovirus in medulloblastomas reveals a potential therapeutic target. J Clin Investig. 2011;121:4043–55. doi: 10.1172/JCI57147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehring GC, Shen HM, Jensen HM, et al. Exposure to bovine leukemia virus is associated with breast cancer: a case–control study. PLoS One. 2015;10:e013–4304. doi: 10.1371/journal.pone.0134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai ZZ, Xu JG, Zhou YH, et al. Human cytomegalovirusencoded US28 may act as a tumor promoter in colorectal cancer. World J Gastroenterol. 2016;22:2789–98. doi: 10.3748/wjg.v22.i9.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo JP, Kowalik TF. Human cytomegalovirus immediate early proteins and cell growth control. Gene. 2002;290:19–34. doi: 10.1016/s0378-1119(02)00566-8. [DOI] [PubMed] [Google Scholar]
- Chang WL, Barry PA, Szubin R, et al. Human cytomegalovirus suppresses type I interferon secretion by plasmacytoid dendritic cells through its interleukin 10 homolog. Virology. 2009;390:330–7. doi: 10.1016/j.virol.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbs CS, Soroceanu L, Denham S, et al. Modulation of oncogenic phenotype in human glioma cells by cytomegalovirus IE1-mediated mitogenicity. Cancer Res. 2008;68:724–30. doi: 10.1158/0008-5472.CAN-07-2291. [DOI] [PubMed] [Google Scholar]
- Dziurzynski K, Wei J, Qiao W, et al. Glioma-associated cytomegalovirus mediates subversion of the monocyte lineage to a tumor propagating phenotype. Clin Cancer Res. 2011;17:4642–9. doi: 10.1158/1078-0432.CCR-11-0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shinawi M, Mohamed HT, El-Ghonaimy EA, et al. Human cytomegalovirus infection enhances NF-κB/p65 signaling in inflammatory breast cancer patients. PLoS One. 2013;8:e55755. doi: 10.1371/journal.pone.0055755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawzy S, Sallam M, Awad NM. Detection of Epstein–Barr virus in breast carcinoma in Egyptian women. Clin Biochem. 2008;41:486–92. doi: 10.1016/j.clinbiochem.2007.12.017. [DOI] [PubMed] [Google Scholar]
- Goodrum F, Caviness K, Zagallo P. Human cytomegalovirus persistence. Cell Microbiol. 2012;14:644–55. doi: 10.1111/j.1462-5822.2012.01774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Handfield J, Toma E, et al. Human cytomegalovirus glycoprotein B genotypes in blood of AIDS patients: lack of association with either the viral DNA load in leukocytes or presence of retinitis. J Med Virol. 1999;59:98–103. [PubMed] [Google Scholar]
- Habibian A, Makvandi M, Samarbafzadeh A, et al. Epstein-Barr Virus DNA frequency in paraffin embedded tissues of Non-Hodgkin lymphoma patients from Ahvaz, Iran. Jundishapur J Health Res. 2013;4:315–20. [Google Scholar]
- Hachana M, Amara K, Ziadi S, et al. Investigation of Epstein–Barr virus in breast carcinomas in Tunisia. Pathol Res Pract. 2011;207:695–700. doi: 10.1016/j.prp.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Hamide M, Makvandi M, Alireza S, et al. Association of human cytomegalovirus with Hodgkin’s disease and non-Hodgkin’s lymphomas. Asian Pac J Cancer Prev. 2017;18:593–7. doi: 10.22034/APJCP.2017.18.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkins LE, Matlaf LA, Soroceanu L, et al. Detection of human cytom- egalovirus in normal and neoplastic breast epithelium. Herpesviridae. 2010;1:8. doi: 10.1186/2042-4280-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwardt T, Lukas S, Zenger M, et al. Human cytomegalovirus immediate-early 1 protein rewires upstream STAT3 to downstream STAT1 signaling switching an IL6-type to an IFNγ-like response. PLoS Pathog. 2016;12:e1005748. doi: 10.1371/journal.ppat.1005748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng B, Glenn W, Ye Y, et al. Human papilloma virus is associated with breast cancer. Br J Cancer. 2009;101:1345–50. doi: 10.1038/sj.bjc.6605282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Jones BC, Logsdon NJ, Josephson K, et al. Crystalstructure of human cytomegalovirus IL-10 bound to soluble human IL-10R1. Proc Natl Acad Sci U S A. 2002;99:9404–9. doi: 10.1073/pnas.152147499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi D, Quadri M, Gangane N, et al. Association of Epstein Barr virus infection (EBV) with breast cancer in rural Indian women. PLoS One. 2009;4:e8180. doi: 10.1371/journal.pone.0008180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Hosseini SZ, Nikkhoo B, et al. Relative frequency of Cytomegalovirus (CMV) in tissue samples of women with breast cancer in Sanandaj. Iran Int J Bioassays. 2016;5:4907–11. [Google Scholar]
- Libard S, Popova SN, Amini RM, et al. Human cytomegalovirus tegument protein pp65 is detected in allintra- and extra- axial brain tumours independent of the tumour type or grade. PLoS One. 2014;9:e108861. doi: 10.1371/journal.pone.0108861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas KG, Bao L, Bruggeman R, et al. The detection of CMV pp65 and IE1 in glioblastoma multiforme. J Neurooncol. 2011;103:231–8. doi: 10.1007/s11060-010-0383-6. [DOI] [PubMed] [Google Scholar]
- Maussang D, Langemeijer E, Fitzsimons CP, et al. The human cytomegalovirus-encoded chemokine receptor US28 promotes angiogenesis and tumor formation via cyclooxygenase-2. Cancer Res. 2009;69:2861–9. doi: 10.1158/0008-5472.CAN-08-2487. [DOI] [PubMed] [Google Scholar]
- Melana SM, Nepomnaschy I, Hasa J, et al. Detection of human mammary tumor virus proteins inhuman breast cancer cells. J Virol Methods. 2010;163:157–61. doi: 10.1016/j.jviromet.2009.09.015. [DOI] [PubMed] [Google Scholar]
- Michaelis M, Doerr HW, Cinatl J. The story of human cytomegalovirus and cancer: increasing evidence and open questions. Neoplasia. 2009;11:1–9. doi: 10.1593/neo.81178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadizadeh F, Zarean M, Abbasi M. Association of Epstein-Barr virus with invasive breast carcinoma and its impact on well-known clinic- pathologic parameters in Iranian women. Adv Biomed Res. 2014;3:141. doi: 10.4103/2277-9175.135158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadizadeh F, Mahmudi F. Evaluation of human cytomegalo- virus antigen expression in invasive breast carcinoma in a population of Iranian patients. Infect Agents Cancer. 2017;12:39. doi: 10.1186/s13027-017-0148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montag C, Wagner JA, Gruska I, et al. The latency associated UL138 gene product of human cytomegalovirus sensitizes cells to tumor necrosis factor alpha (TNF-α) signaling by upregulating TNF-α receptor 1 cell surface expression. J Virol. 2011;85:11409–21. doi: 10.1128/JVI.05028-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrucelli A, Umashankar M, Zagallo P, et al. Interactions between proteins encoded within the human cytomegalovirus UL133-UL138 locus. J Virol. 2012;86:8653–62. doi: 10.1128/JVI.00465-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AK, Currie MJ, Robinson BA, et al. Cytomegalovirus and Epstein-Barr virus in breast cancer. PLoS One. 2015;10:e0118989. doi: 10.1371/journal.pone.0118989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AK, Cox B, McCredie MR, et al. Cytomegalovirus, Epstein–Barr virus and risk of breast cancer before age 40 years: a case–control study. Br J Cancer. 2004;90:2149–52. doi: 10.1038/sj.bjc.6601822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadjadi A, Nouraie M, Ghorbani A, et al. Epidemiology of breast cancer in the Islamic Republic of Iran first results from a population-based cancer registry. East Mediterr Health J. 2009;15:1426–31. [PubMed] [Google Scholar]
- Samanta M, Harkins L, Klemm K, et al. High prevalence of human cytomegalovirus in prostatic intraepithelial neoplasia and prostatic carcinoma. J Urol. 2003;170:998–1002. doi: 10.1097/01.ju.0000080263.46164.97. [DOI] [PubMed] [Google Scholar]
- Sanchez V, Spector DH. Subversion of cell cycle regulatory pathways. Curr Top Microbiol Immunol. 2008;325:243–62. doi: 10.1007/978-3-540-77349-8_14. [DOI] [PubMed] [Google Scholar]
- Shahab M, Akbar S, Nasrollah E, et al. Presence of human Papilloma virus DNA in colorectal cancer tissues in Shiraz, Southwest Iran. Asian Pac J Cancer Prev. 2015;16:7883–87. doi: 10.7314/apjcp.2015.16.17.7883. [DOI] [PubMed] [Google Scholar]
- Slinger E, Maussang D, Schreiber A, et al. HCMV-encoded chemokine receptor US28 mediates proliferative signaling through the IL-6-STAT3 axis. Sci Signal. 2010;3:ra58. doi: 10.1126/scisignal.2001180. [DOI] [PubMed] [Google Scholar]
- Slobedman B, Barry PA, Spencer JV, et al. Virus-encoded homologs of cellular interleukin-10 and their control of host immune function. J Virol. 2009;83:9618–29. doi: 10.1128/JVI.01098-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soroceanu L, Matlaf L, Bezrookove V, et al. Human cytomegalovirus US28 found in glioblastoma promotes an invasive and angiogenic phenotype. Cancer Res. 2011;71:6643–53. doi: 10.1158/0008-5472.CAN-11-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafvizi F, Fard ZT. Detection of human cytomegalovirusin patients with colorectal cancer by nested-PCR. Asian PacJ Cancer Prev. 2014;15:1453–7. doi: 10.7314/apjcp.2014.15.3.1453. [DOI] [PubMed] [Google Scholar]
- Taher C, de Boniface J, Mohammad AA, et al. High prevalence of human cytomegalovirus proteins and nucleic acids in primary breast cancer and metastatic sentinel lymph nodes. PLoS One. 2013;8:e56795. doi: 10.1371/journal.pone.0056795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utrera-Barillas D, Valdez-Salazar H-A, Gómez-Rangel D, et al. Is human cytomegalovirus associated with breast cancer progression? Infect Agents Cancer. 2013;8:12. doi: 10.1186/1750-9378-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Liu P, Zhang B, et al. Role of STAT3 decoy oligodeoxy nucleotides on cell invasion and chemo sensitivity in human epithelial ovarian cancer cells. Cancer Genet Cytogenet. 2010;197:46–53. doi: 10.1016/j.cancergencyto.2009.10.004. [DOI] [PubMed] [Google Scholar]


