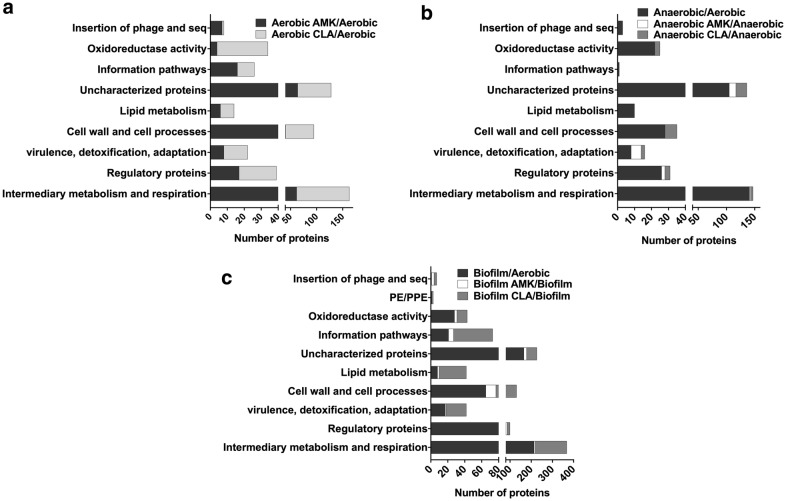
Fig. 5.
Functional classification of MAH proteins synthesized 1.5-fold and more was done by finding the protein homologs of H37Rv strain of Mycobacterium tuberculosis and using the functional categorization available on TubercuList webserver of Institute Pasteur. MAH proteins that did not match to any proteins of H37Rv strain were classified based on predicted or conserved domain/motif search at NCBI (National Center for Biotechnology Information) and the Pfam database through the Sanger Institute. Histograms show number of proteins belonging to (a) aerobic, (b) anaerobic or (c) biofilm functional category with and without antibiotic treatment