Lung cancer is the most fatal malignancy in China. This study assessed genomic alterations of driver genes in a cohort of Chinese patients with non‐small cell lung cancer. This article reports the resulting analysis of germline mutations, EGFR variations, and ALK rearrangements, the most common and important driver genes in Chinese NSCLC population.
Keywords: Non‐small cell lung cancer, Next‐generation sequencing, Molecular genomic profile
Abstract
Background.
Non‐small cell lung cancer (NSCLC) is one of the most common human malignancies and the leading cause of cancer‐related death. Over the past few decades, genomic alterations of cancer driver genes have been identified in NSCLC, and molecular testing and targeted therapies have become standard care for lung cancer patients. Here we studied the unique genomic profile of driver genes in Chinese patients with NSCLC by next‐generation sequencing (NGS) assay.
Materials and Methods.
A total of 1,200 Chinese patients with NSCLC were enrolled in this study. The median age was 60 years (range: 26–89), and 83% cases were adenocarcinoma. NGS‐based genomic profiling of major lung cancer‐related genes was performed on formalin‐fixed paraffin‐embedded tumor samples and matched blood.
Results.
Approximately 73.9% of patients with NSCLC harbored at least one actionable alteration recommended by the National Comprehensive Cancer Network guideline, including epidermal growth factor receptor (EGFR), ALK, ERBB2, MET, BRAF, RET, and ROS1. Twenty‐seven patients (2.2%) harbored inherited germline mutations of cancer susceptibility genes. The frequencies of EGFR genomic alterations (both mutations and amplification) and ALK rearrangement were identified as 50.1% and 7.8% in Chinese NSCLC populations, respectively, and significantly higher than the Western population. Fifty‐six distinct uncommon EGFR mutations other than L858R, exon19del, exon20ins, or T790M were identified in 18.9% of patients with EGFR‐mutant NSCLC. About 7.4% of patients harbored both sensitizing and uncommon mutations, and 11.6% of patients harbored only uncommon EGFR mutations. The uncommon EGFR mutations more frequently combined with the genomic alterations of ALK, CDKN2A, NTRK3, TSC2, and KRAS. In patients <40 years of age, the ALK‐positive percentage was up to 28.2%. Moreover, 3.2% of ALK‐positive patients harbored multi ALK rearrangements, and seven new partner genes were identified.
Conclusion.
More unique features of cancer driver genes in Chinese NSCLC were identified by next‐generation sequencing. These findings highlighted that NGS technology is more feasible and necessary than other molecular testing methods, and suggested that the special strategies are needed for drug development and targeted therapy for Chinese patients with NSCLC.
Implications for Practice.
Molecular targeted therapy is now the standard first‐line treatment for patients with advanced non‐small cell lung cancer (NSCLC). Samples of 1,200 Chinese patients with NSCLC were analyzed through next‐generation sequencing to characterize the unique feature of uncommon EGFR mutations and ALK fusion. The results showed that 7.4% of EGFR‐mutant patients harbored both sensitizing and uncommon mutations and 11.6% harbored only uncommon mutations. Uncommon EGFR mutations more frequently combined with the genomic alterations of ALK, CDKN2A, NTRK3, TSC2, and KRAS. ALK fusion was more common in younger patients, and the frequency decreased monotonically with age. 3.2% of ALK‐positive patients harbored multi ALK rearrangement, and seven new partner genes were identified.
Introduction
Lung cancer is the most fatal malignancy in China [1], [2]. An estimated 733,300 newly diagnosed lung cancer cases and 610,200 lung cancer‐related deaths were reported in China in 2015, and traditional surgery and radio‐chemotherapy are of limited help to improve the survival of the patients [3]. Following the discovery of cancer driver genes and the development of next‐generation sequencing (NGS) technology, genomic alterations have been integrated as part of the standard diagnostic procedure, and several targeted drugs targeting the driver gene mutations have been applied in clinic and achieved certain therapeutic effects [4], [5], [6], [7].
The major type of lung cancer is non‐small cell lung cancer (NSCLC), which accounts for approximately 85% of all cases [8]. Epidermal growth factor receptor (EGFR) gene mutations can be detected in approximately 10%–20% in Western patients with advanced NSCLC, but as high as 40%–50% in the nonsmoking Asian population [9], [10], [11]. In 2003, EGFR alterations, including point mutations and short insertions/deletions (indels), were identified as the first druggable mutations in NSCLC and proved to be the most potent predictive biomarker for EGFR tyrosine kinase inhibitors (TKIs) [7]. Thus far, the U.S. Food and Drug Administration (FDA) has approved several EGFR TKIs for NSCLC. Additionally, the Chinese National Medical Products Administration has approved the first‐generation EGFR‐TKI, icotinib, for first‐line treatment of patients with metastatic NSCLC with sensitizing EGFR mutations [12], [13], [14], [15], [16]. In NSCLC, the most common sensitizing EGFR alterations are deletions in exon 19 (approximately 45% of patients with EGFR alterations) and L858R mutation in exon 21 (approximately 40%) [10]. But more than half of patients with disease progression after response to initial EGFR‐TKI treatment harbor an acquired TKI‐resistant EGFR T790 M mutation [17], [18], [19]. The third‐generation EGFR TKI, osimertinib, developed to target EGFR T790 M and other EGFR sensitizing mutations, has been approved by the FDA for the first‐line treatment of patients with metastatic NSCLC with EGFR exon 19del or L858R and the treatment of patients with metastatic EGFR T790 M mutation‐positive NSCLC whose disease has progressed on or after EGFR‐TKI therapy [20]. In addition, first‐generation EGFR TKI is effective in treating NSCLC with common EGFR sensitizing mutations (exon 19deletion and L858R), but it is less effective in treating NSCLC with uncommon EGFR mutations, such as G719X or L861Q [21]. The average progression‐free survival (PFS) of patients with nonconventional EGFR‐carrying lung adenocarcinoma treated with gefitinib is only 7.7 months, significantly (p < .001) shorter than the 11.4 months of patients with common EGFR‐carrying lung adenocarcinoma also treated with gefitinib [22]. However, according to the combined analysis of LUX‐Lung 2, LUX‐Lung 3, and LUX‐Lung 6, the second‐generation TKI afatinib was active in patients with NSCLC who harbored certain types of uncommon EGFR mutations, especially G719, L861Q, and S768I [23].
Gene fusion or rearrangement, especially kinase gene fusion, is another typical genomic variation in lung cancer. The most common rearrangement genes in NSCLC are ALK [24], [25], ROS1 [26], and RET [27], representing 2%–7%, 1%–2% and 1%–2% of patients, respectively [25], [26], [27], [28], [29], [30]. The echinoderm microtubule‐associated protein‐like 4 (EML4)‐ALK constitutes a major subset of ALK fusions. Patients with NSCLC with EML4‐ALK likely are of adenocarcinoma histology, never/light smokers, men, and younger, and they do not benefit from EGFR TKIs [31]. Similarly, ROS1 rearrangements occur more frequently in younger women with adenocarcinoma histology and never smokers [26], [32]. Patients with ALK and ROS1 rearrangements are predicted to benefit from TKIs, such as crizotinib [33], [34], [35], [36]. The National Comprehensive Cancer Network (NCCN) Guideline for NSCLC also recommends cabozantinib and vandetanib for patients with RET rearrangements [37], [38], [39].
In the present study, genomic alterations of driver genes were assessed in a Chinese NSCLC cohort of 1,200 patients by NGS in the College of American Pathologists (CAP)‐accredited lab. We described the basic profile of the patient's gene mutations, particularly those that are targeted therapy available, and compared them with The Cancer Genome Atlas (TCGA) and published data (mainly Western people in Europe and the U.S.). Germline mutations can be passed on to offspring to form hereditary cancers. Thus, we also analyzed the germline mutation in Chinese patients with NSCLC. Finally, we analyzed the unique pattern of EGFR variations and ALK rearrangements, the most common and important driver genes in Chinese NSCLC populations.
Materials and Methods
Samples Source and Ethic Data
In total, 1,200 formalin‐fixed, paraffin‐embedded (FFPE) tumor samples and matched blood samples were collected and prepared according to standard procedures. All the cases were diagnosed as NSCLC according to World Health Organization criteria based on hematoxylin and eosin staining reviewed by experienced pathologists. All subjects involved in this study were clearly informed, understood the contents of the study, and agreed to publish the results of the study. All patients had signed an informed consent.
Next‐Generation Sequencing
Genomic profiling was performed in the laboratory at OrigiMed (Shanghai, China), a CAP‐accredited lab. At least 50 ng of cancer tissue DNA was extracted from each 40 mm FFPE tumor sample using a DNA Extraction Kit (QIAamp DNA FFPE Tissue Kit; Qiagen, Hilden, Germany) according to manufacturer's protocols. All the coding exons of 37 key cancer‐related genes and selected introns of 8 genes (supplemental online Table 1) were captured by the custom hybridization capture panel. In addition, the probe density was increased to ensure high efficiency of capture in the conservatively low read depths region. Libraries were each diluted to 1.05 nM and then sequenced with a mean coverage of 900× for FFPE samples (minimum 700×) and 300× for matched blood samples on an Illumina NextSeq‐500 Platform (Illumina Incorporated, San Diego, CA).
Bioinformatics Analysis
Genomic alterations, including single nucleotide variants (SNVs), short and long insertions/deletions (indels), copy number variations (CNVs), and gene rearrangements, were subjected to advanced analysis. First, reads were aligned to human genome reference sequence (hg19) by Burrows‐Wheeler Aligner, and polymerase chain reaction (PCR) duplicates were removed using Picard. Second, SNVs and short indels were identified by MuTect after quality recalibration and realignment using Genome Analysis Toolkit (GATK) and in‐house pipeline. Short indels were then calibrated using the results from Pindel. Read depths were normalized within target regions by Exome Copy number Alterations/Variations annotATOR (EXCATOR). The log‐ratio per region of each gene was calculated, and customized algorithms were used to detect copy number changes. Tumor cellularity was estimated by allele frequencies of sequenced SNPs. A customized algorithm was developed to detect gene rearrangements and long indels.
Reliable somatic alterations were detected in the raw data by comparison with matched blood control samples. At minimum, five reads and minimum variant allele frequency of 1% were required to support alternative calling. For CNVs, focal amplifications were characterized as genes with thresholds ≥4 copies for amplification and 0 copies for homozygous deletions. For the calling of gene rearrangements, aligned reads with abnormal insert size of over 2,000 or zero bp were collected and used as discordant reads. Next, the discordant reads with the distance less than 500 bp formed clusters that were further assembled to identify potential rearrangement breakpoints. The breakpoints were reconfirmed by the BLAST‐like alignment tool and the resulted chimeric gene candidates were annotated. Clinically relevant genomic alterations were further marked as druggable genomic alterations in current treatments or clinical trials. IBM SPSS Statistics (Version 20.0; IBM Corp, Armonk, NY) was used for statistical analysis. For all test, p < .05 was defined as statistically significant.
Results
Demographic and Clinicopathological Data of the Patients
The demographics and clinicopathological data of patients in the cohort are summarized in Table 1. The median age of patients at the time of sampling was approximately 60 years (range: 26–89 years), and males were moderately overrepresented compared with females (56% of patients were male). Although there were approximately equal numbers of male and female patients younger than 60, male patients were significantly overrepresented in the ≥60 age group (61% vs. 39%, p = .0007). Regarding histological subtypes, adenocarcinoma was the most common subtype (83%) and large cell carcinoma was the least common subtype (0.8%). In addition, .9% of the cases were of uncertain subtypes. Patients were classified into main clinical stages (I–IV) according to both pathology and medical history following the American Journal of Critical Care Cancer Staging Manual (version eight; Table 1). Patients in different stages had similar age distributions (the median was approximately 60 years of age). Males were particularly prevalent in the older (≥60 years of age) and late‐stage (III–IV) groups.
Table 1. Clinical characteristics of 1,200 Chinese patients with NSCLC.
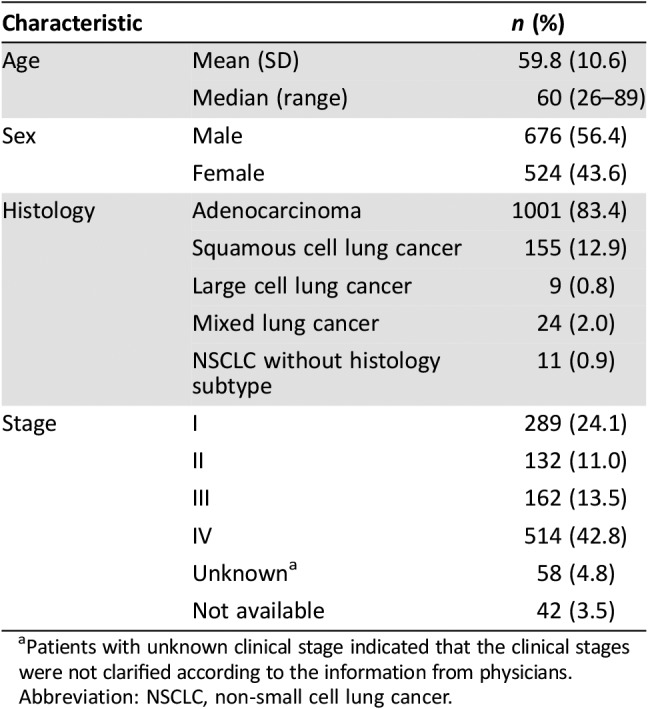
Patients with unknown clinical stage indicated that the clinical stages were not clarified according to the information from physicians.
Abbreviation: NSCLC, non‐small cell lung cancer.
Common and Uncommon EGFR Alterations in Chinese Patients with NSCLC
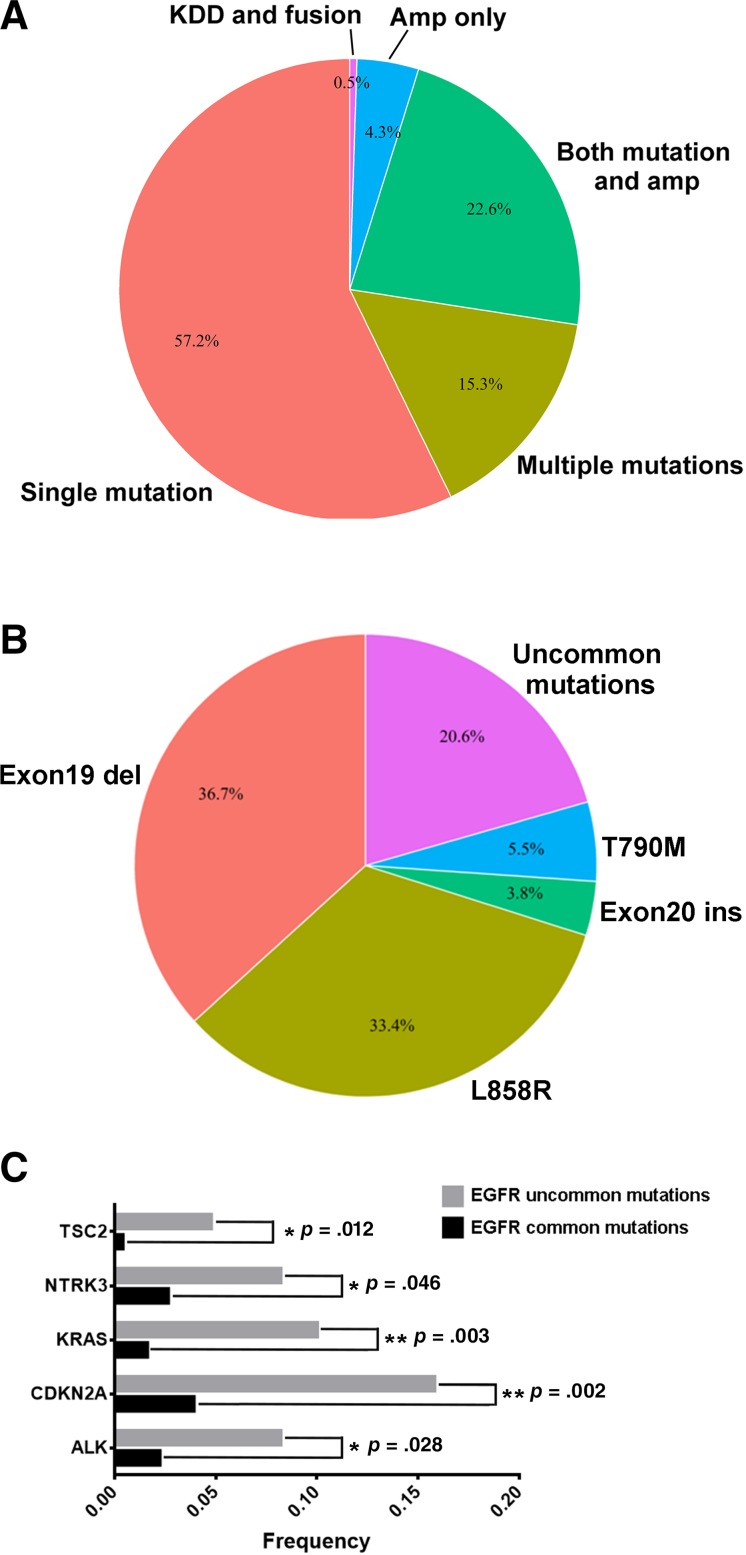
In total, 47.6% of the 1,200 patients were identified to have EGFR mutations, which excluded amplification of EGFR. 88.4% of EGFR‐mutant patients harbored hotspots, including L858R (33.4%), exon 19 deletion (36.7%), T790M (5.5%), and exon 20 insertions (3.8%). Figure 1A showed the profile of co‐occurring EGFR genomic alterations. The majority (57.2%) possessed only mutations (SNV and indel), and a decent number had both mutations and amplifications (22.6%); 4.8% had only amplifications (4.3%) and structure variations (0.5%) including kinase domain duplication (KDD) and EGFR‐RAD51 rearrangement. The percentage of patients with NSCLC with multiple EGFR mutations was 15.3% of the patients with EGFR‐altered NSCLC. Moreover, uncommon mutations accounted for 20.6% of EGFR mutations (Fig. 1B).
Figure 1.
Distribution of EGFR mutations in Chinese cohort. Profiling of overlapping EGFR alteration types (A) and mutation spectra among Chinese patients with EGFR‐mutant non‐small cell lung carcinoma (B). Co‐occurring of EGFR common and uncommon mutations with other genomic alterations were analyzed (C). *Indicates p < .05; **Indicates p < .01.
Abbreviations: EGFR, epidermal growth factor receptor; KDD, kinase domain duplication.
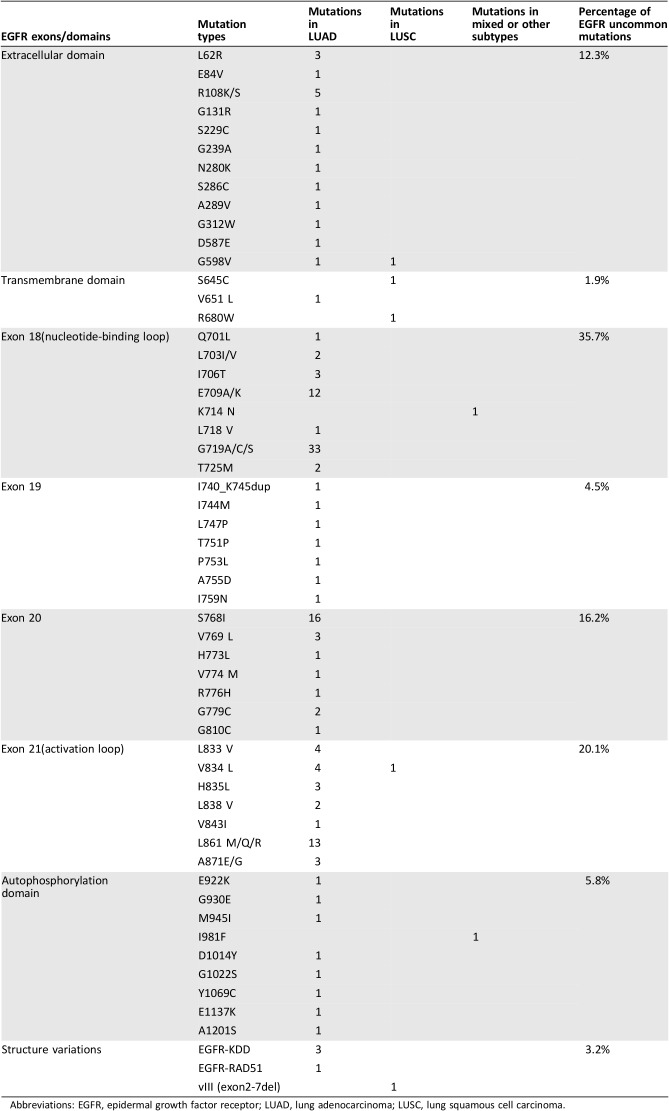
Uncommon EGFR mutations were defined as the mutations other than L858R, exon 19del, exon 20ins, and T790M. In our study, 154 uncommon EGFR mutations in 108 patients, which accounted for 18.9% of EGFR‐mutant patients, were identified. Two previous studies reported that approximately 6.5% of uncommon EGFR‐mutant patients were detected in Chinese patients with NSCLC by Sanger sequencing or amplification refractory mutation system (ARMS) technologies [40], [41]. Uncommon EGFR mutations that co‐occurred with sensitizing mutations were identified in 7.4% of EGFR‐mutant patients, and 11.6% of EGFR‐mutant patients harbored only uncommon mutations. Among all the uncommon mutations, 12.3% occurred in the extracellular domain, 1.9% were in the transmembrane domain, 5.8% were in the autophosphorylation domain, 76.6% were in the kinase domain of exon 18–21, and 3.2% were structure variations of KDD and rearrangements (Table 2). A total of 35.7% of the uncommon mutations occurred in exon 18. G719X, S768I, L861X, and E709X were the most frequently identified recurrent uncommon mutations, which accounted for 21.4%, 10.4%, 8.4%, and 7.8% of uncommon EGFR mutations, respectively.
Table 2. Uncommon EGFR mutations identified in Chinese patients with non‐small cell lung cancer.
Abbreviations: EGFR, epidermal growth factor receptor; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
EGFR alterations occurred more frequently in females (in contrast to all other driver gene mutations) and often in combination with TP53 mutations; patients with EGFR‐mutated squamous carcinoma were mainly males (82%) and always carried TP53 comutations. The age, sex, and histological type characteristics of patients with NSCLC with only uncommon EGFR mutations or common mutations are summarized in Table 3. The incidence of uncommon EGFR mutations only (4.5%) was much higher than that of common EGFR mutations (0.6%) in lung squamous cancer.
Table 3. Comparison of characteristics between common and uncommon EGFR mutations in Chinese NSCLC patients.
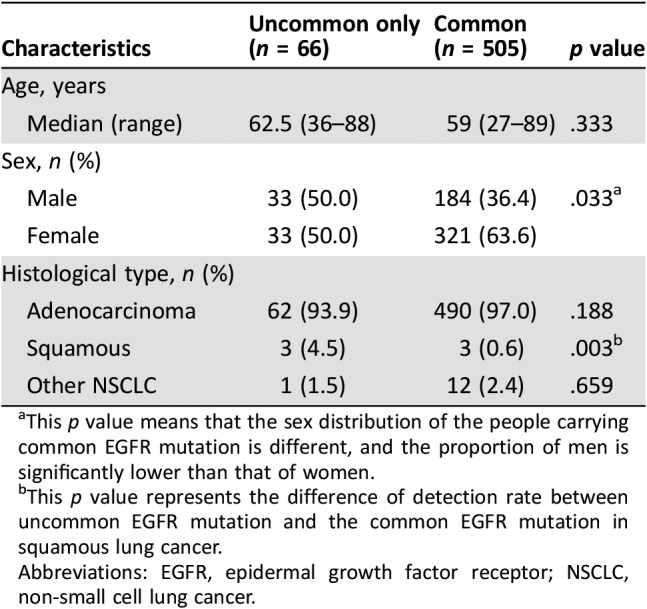
This p value means that the sex distribution of the people carrying common EGFR mutation is different, and the proportion of men is significantly lower than that of women.
This p value represents the difference of detection rate between uncommon EGFR mutation and the common EGFR mutation in squamous lung cancer.
Abbreviations: EGFR, epidermal growth factor receptor; NSCLC, non‐small cell lung cancer.
Uncommon EGFR mutations were found to occur combined with genomic alterations of CDKN2A (p = .002), KRAS (p = .003), TSC2 (p = .012), ALK (p = .028), and NTRK3 (p = .046) more frequently (Fig. 1C). Further analysis of co‐occurring EGFR mutations and kinase receptor fusions revealed that six Chinese patients with EGFR‐mutant NSCLC harbored both EGFR mutations and known druggable kinase receptor rearrangements, including ROS1, RET, and NTRK. Three of the six patients received EGFR TKI as the standard treatment. One patient achieved partial response for 10 months, and two patients achieved stable disease for 5 and 4 months.
ALK Rearrangement in Chinese Patients with NSCLC
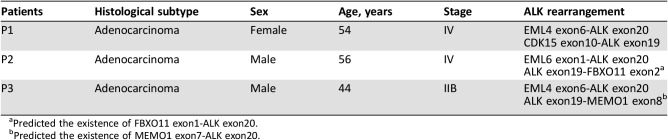
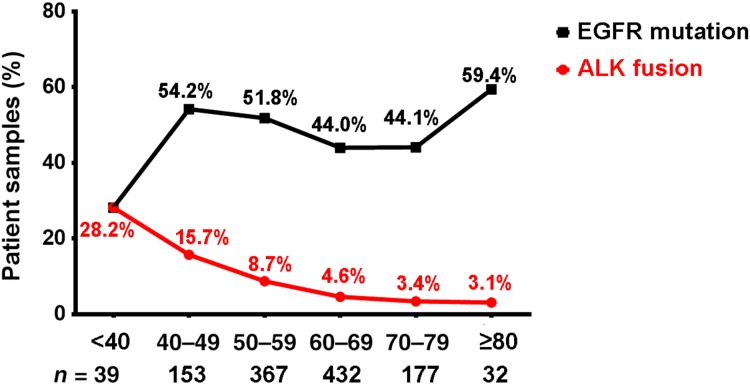
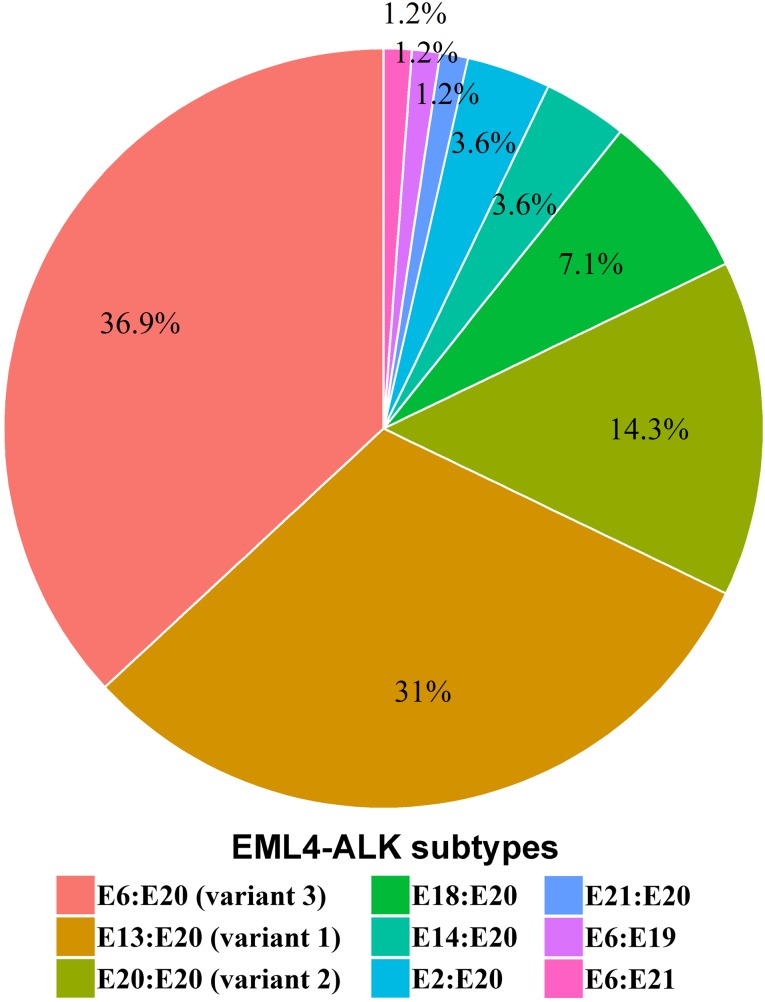
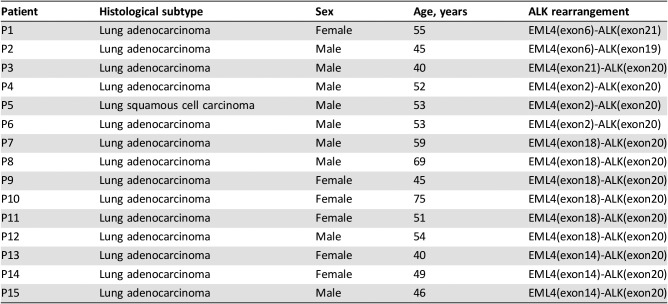
In the Chinese cohort, we identified 97 ALK rearrangements in 94 patients, and there were 3 patients (3.2%) who harbored multi ALK rearrangements (Table 4). Younger patients were more likely to harbor ALK rearrangements (Fig. 2). In patients aged <40 years, the ALK‐positive percentage was up to 28.2%, and the percentage decreased along with age to 3.1% in patients aged 70 years and above. The number of Chinese patients with NSCLC with age <40 was relatively low (n = 39). One in three of such younger patients harbored ALK rearrangements and may potentially benefit from targeted ALK‐inhibitor therapies. In terms of gender, the ALK‐positive percentages in male and female patients with NSCLC were 7.2% and 8.6%, respectively, but the difference was not statistically significant. Analysis of the partner genes of ALK rearrangement identified in Chinese patients with NSCLC revealed that EML4‐ALK contributed to 88.7% of ALK rearrangement, and seven new partners were identified in our study including CDK15‐ALK, EML6‐ALK, FBXO11‐ALK, CAMKMT‐ALK, YAP1‐ALK, MEMO1‐ALK, and LCLAT1‐ALK. The frequency of EML4‐ALK subtypes is shown in Figure 3. The most common subtypes of EML4‐ALK were E6:E20 (variant 3; 36.9% of all EML4‐ALK cases) and E13:E20 (variant 1; 31.0% of all EML4‐ALK cases), whereas E20:E20 (variant 2) accounted for 14.3% of all EML4‐ALK cases. Interestingly, E6‐E20 was enriched in male patients (18 vs. 13), whereas E13‐E20 was enriched in female patients (15 vs. 11). Other uncommon subtypes of EML4‐ALK in the Chinese cohort were identified (Table 5).
Table 4. Patient characteristics with dual ALK rearrangements.
Predicted the existence of FBXO11 exon1‐ALK exon20.
Predicted the existence of MEMO1 exon7‐ALK exon20.
Figure 2.
The incidence of ALK fusion and EGFR mutations in Chinese NSCLC according to different age groups. Red curve represents the frequency of patients with ALK fusion in each age group. Black curve represents the frequency of patients with EGFR mutations in each age group.
Figure 3.
The frequency and distribution of EML4‐ALK fusion subtypes identified in Chinese NSCLC cohort.
Table 5. Patient characteristics with uncommon EML4‐ALK rearrangement.
Profiling of Genomic Alterations in 1,200 Chinese Patients with NSCLC
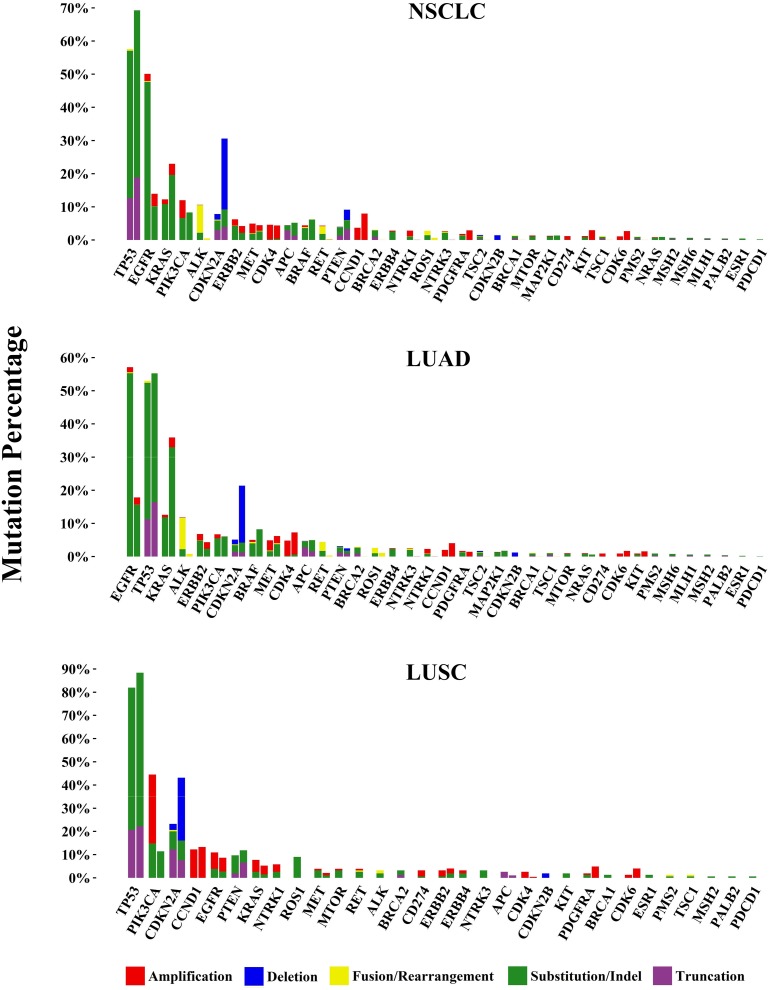
Overall, the average mutations per cancer sample were 2.5 and 2.3 in the samples from male and female patients, respectively. The incidence of genomic alteration is summarized in Figure 4. For each gene, the mutational profile (frequency and composition) in the Chinese cohort (left column) was compared with that in the corresponding TCGA data sets (right column) [42], which were mostly based on Western populations and whole exome sequencing. The genomic alteration incidences of most of the 37 lung cancer‐related genes showed statistic differences between Chinese and Western not only in total NSCLC populations but also the subtypes of adenocarcinoma and squamous cell carcinoma. Different genes had markedly distinct composition of alteration types. For example, TP53 and EGFR were the most frequently mutated genes, and both had SNV as the major alteration type. Of particular interest were common actionable genomic alterations (involving EGFR, KRAS, ALK, ROS1, RET, MET, BRAF, PIK3CA, PTEN, and NTRK1/3) in the Chinese NSCLC cohort. The most frequent genomic alterations were EGFR (50.1% of cases harbored at least one EGFR mutation or amplification), KRAS (12.3% of cases harbored KRAS mutations or amplification), and ALK rearrangement (7.8%). Other frequent genomic alterations were as follows: PIK3CA (12.0% both for mutations or amplification), ERBB2 alterations (6.3%; 4.3% mutations and 2.4% amplifications), PTEN (4.0% both for mutations or deletion), BRAF (4.4%), MET alterations (3.4%; 3.0% amplifications and 0.4% exon 14 skipping mutations), RET rearrangement (2.3%), and ROS1 rearrangement (1.3%).
Figure 4.
Genomic alterations in 1,200 cases of Chinese non‐small cell lung carcinoma. 37 lung cancer‐related genes and their composition of alteration types, including clinically relevant genomic alterations of the driver genes, are shown by cohort as a whole (NSCLC) as well as by adenocarcinoma cases (LUAD) and squamous carcinoma cases (LUSC). For comparison, corresponding The Cancer Genome Atlas figures are juxtaposed on the right.
Abbreviations: LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; NSCLC, non‐small cell lung cancer.
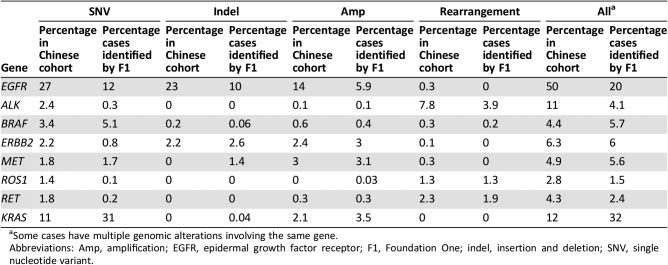
Approximately 73.9% of cases harbored at least one genomic alteration, including SNV, short indel, long indel, CNV, and gene rearrangement, in druggable genes recommended by the NCCN guideline, including EGFR, ALK, ERBB2, MET, BRAF, RET, and ROS1, which showed ethnic differences between Chinese and Western NSCLC populations identified by target sequencing assay of Foundation One as well [9] (Table 6). Gene rearrangements in kinase genes including ALK, BRAF, EGFR, NTRK1/3, MET, ERBB2, PDGFRA, RET, and ROS1 occurred in 13.2% of Chinese patients with NSCLC. MET exon 14 skipping was detected at 0.4% compared with that commonly previously reported at 3%, which may represent a difference in Chinese versus Western populations.
Table 6. Comparison of genomic alterations involving National Comprehensive Cancer Network guideline genes and KRAS between Chinese and Western non‐small cell lung carcinoma.
Some cases have multiple genomic alterations involving the same gene.
Abbreviations: Amp, amplification; EGFR, epidermal growth factor receptor; F1, Foundation One; indel, insertion and deletion; SNV, single nucleotide variant.
In addition to the common genes mentioned above, the incidences of some other less common genes were also compared in our study. Potential actionable tumor suppressor genes such as CDKN2A/B, BRCA1/2, PTEN, and TSC1/2 showed statistic differences in prevalence between Chinese and Western populations with NSCLC. Considering the histological subtypes of NSCLC, lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) displayed different patterns of racial diversity. The frequency of cell cycle‐related genes CDKN2A was much lower in both LUAD (5.1% vs. 21.5%, p < .0001) and LUSC (23.2% vs. 43.6%, p = .0024), and genomic alterations of BRCA2 occurred more in the Chinese population than the Western cohort in both LUAD (2.9% vs. 0.0%, p < .0001) and LUSC (3.2% vs. 0.0%, p = .0008). However, the prevalence of TSC1/2 and BRCA1 only showed difference in LUAD and not in LUSC.
Germline Mutation of Cancer Susceptibility Genes in Chinese Patients with NSCLC
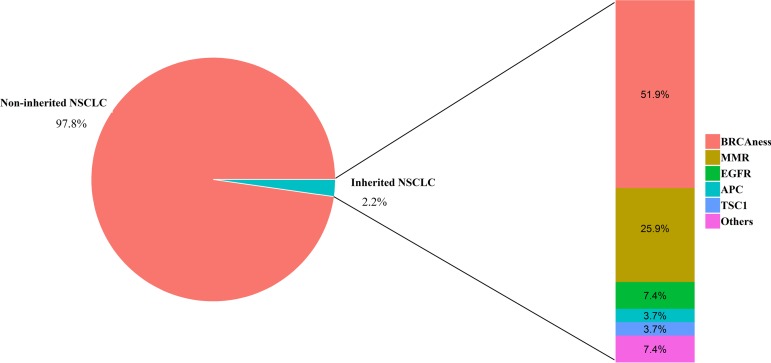
In the present study, 27 patients (2.2%) with NSCLC harbored inherited germline mutations of cancer susceptibility genes (Fig. 5). Of those, the most frequently occurring were BRCAness genes (14 patients, 51.9%). BRCAness is a term used for genes whose mutations in cancer cells behave similarly to tumors with BRCA1 or BRCA2 mutations. Loss of function germline mutations of BRCAness genes included mutations on BRCA1/2 (five patients), FANCA (four patients), ATM (one patient), BARD1 (one patient), BRIP1 (one patient), CHEK2 (one patient), and PALB2 (one patient). In addition, seven patients harbored inactivating germline mutations of mismatch repair (MRR) genes, including MLH1 (one patient), MSH2 (one patient), MSH6 (one patient), PMS2 (three patients), and EPCAM (one patient). The following two EGFR germline mutations were also identified in the Chinese cohort: a T790 M mutation in a 27‐year‐old male adenosquamous subtype patient and a G724S mutation in a 57‐year‐old female adenocarcinoma subtype patient; both patients have a family history of lung cancer. To the best of our knowledge, this is the first report of the germline EGFR G724S mutation. Previous studies have suggested that EGFR G724S is oncogenic [43], [44] and that the EGFR T790M germline mutation is associated with inherited NSCLC [45], [46], [47]. Other pathogenic germline mutations were identified in APC, TSC1, SDHC, and SPINK1. In addition, patients with pathogenic germline mutations of cancer susceptibility genes were 7 years younger than patients without those mutations (54 vs. 61 years of age).
Figure 5.
Germline mutations of cancer susceptibility genes identified with a multigene panel among 1,200 Chinese individuals with non‐small cell lung carcinoma.
Abbreviations: EGFR, epidermal growth factor receptor; MMR, mismatch repair; NSCLC, non‐small cell lung cancer.
Discussion
Most patients with NSCLC are diagnosed at the middle to late stage despite recent advances of early detection screening for lung cancer. Fortunately, NGS technology now is in wide use to identify cancer gene mutations and provide molecular basis for appropriate targeted therapy in clinical precision medicine. In this NGS‐based study of Chinese patients with NSCLC, a comprehensive profile of genomic alterations was constructed. In gene mutations of EGFR, KRAS, and ALK, Chinese patients with NSCLC were different from the Western population, as the former had a much higher frequency (47.6% vs. 20.0%) of EGFR mutations and ALK rearrangements (7.8% vs. 4.1%) but a significantly lower frequency (10.8% vs. 32.0%) of KRAS mutation than the latter [9], which was consistent with previous reports [48], [49], [50]. Nearly three quarters of Chinese patients with NSCLC harbored at least one druggable genomic alteration of driver genes recommended by the NCCN guideline, revealing the different clinical characteristics of Chinese patients. However, the TCGA data sets were generated by a different technological platform (Affymetrix SNP6.0 arrays) and bioinformatics suite (e.g., GISTIC for CNV calling and RNAseq for fusion calling).
The patients with NSCLC with EGFR mutation may benefit from treatment using EGFR TKIs. In this study, the two most common EGFR mutations identified in Chinese patients with NSCLC were exon 19 deletion (36.7%) and L858R mutation (33.4%), which are sensitive to EGFR TKIs, and this was consistent with the findings of the study published with Western people as the main subjects [51]. The exon 20 insertions, which were mostly insensitive to EGFR TKIs [52], [53], amounted to 3.8% of EGFR mutations, and the most common resistant mutation, T790M [54], [55], accounted for 5.5% of EGFR mutations. For those 5.5% of patients with primary EGFR T790M mutation, which suggests primary resistance, selecting one of the third‐generation EGFR TKIs as first‐line therapy is advisable. NGS is currently the most effective way to detect EGFR gene alterations [51]. By increasing the depth of sequencing, some less common and low‐frequency mutations may be discovered. In this study, the detection rate of uncommon EGFR mutation only was 11.6% of EGFR mutant patients, which was higher than about 6.5% from previous studies. This finding may be related to the advantage of the whole exon coverage of EGFR of our NGS assay and the increase of sequencing depth. We used 900× depth sequencing and found a total of 56 different types of uncommon EGFR mutations, which could be observed in the extracellular domain (12.3%), transmembrane domain (1.9%), exon 18 (nucleotide‐binding loop, 35.7%), exon 19 (4.5%), exon 20 (16.2%), exon 21 (activation loop, 20.1%), autophosphorylation domain (5.8%), and structure variations (3.2%). Some uncommon EGFR mutations with high incidence (more than 10 cases) included L861 M/Q/R, G719A/C/S, S768I, and E709A/K, which have been described previously [56], [57]. L861Q, G719X, and S768I were EGFR mutations with weak activities; the median PFS of patients who received EGFR TKIs as first‐line treatment was 7.6 months, 8.2 months, and 3.4 months, respectively, which was significantly shorter than that in patients with common EGFR mutations (exon 19 deletion and L858R) [56]. The E709A mutation is located in EGFR exon 18 and often combined with other forms of EGFR mutations and is associated with shorter survival in patients with lung cancer [58]. These results demonstrate that the patients with NSCLC with uncommon EGFR mutations exhibit a significantly inferior PFS compared with those with common EGFR mutations for first‐generation TKIs. However, other studies revealed that the second‐generation TKI afatinib was more active in patients with NSCLC who harbored certain types of uncommon EGFR mutations, especially G719, L861Q, and S768I [23], which indicated that accurate detection of uncommon EGFR mutations would help patients with NSCLC to select targeted therapeutic drugs more precisely. Besides, it was noteworthy that comutation of EGFR and TP53 was frequently found in female patients in this study; another study showed that it was more likely to occur in young patients <45 years of age [57]. At the same time, the common mutation of EGFR and TP53 is correlated with a poor prognosis [59]. It seems that young women should pay more attention to screening for NSCLC because their prognosis is poor in the event of NSCLC.
In addition to common druggable genomic alterations, frequencies of druggable tumor suppressor genes, which were considered uncommon genes in NSCLC, displayed a unique signature in Chinese populations. The incidences of BRCA1/2 and TSC1/2 were higher in the Chinese cohort than in the Western population. BRCA1/2 were the well‐known biomarkers for poly ADP‐ribose polymerase (PARP) inhibitors, and TSC1/2 naturally suppressed the overactivity of downstream mammalian target of rapamycin (mTOR), which indicated the potential clinical benefits of patients with TSC1/2 loss of function mutations from mTOR inhibitors. However, compared with the Western population, the incidence of CDKN2A/B in Chinese patients with NSCLC was lower. Whether loss of function or deletion of CDKN2A/B related with clinical outcome of CDK inhibitors was not clear, but the lower incidence of these two cell cycle‐related genes indicated the differences in pathogenesis in different ethnic groups.
In this Chinese cohort, pathogenic germline mutations were identified in a portion of younger patients, highlighting the necessity of risk assessment and management for those patients and their blood relatives. Two pathogenic or likely pathogenic germline mutations of EGFR, including a novel cancer susceptibility EGFR G724S mutation, were identified.
ALK rearrangement is often seen in NSCLC (3.1%), whereas its incidence in other tumors is only 0.2% [60]. The data in this study showed that the percentage of Chinese patients with ALK rearrangement was 7.8%, quite high compared with the published data mentioned above. Moreover, compared with PCR‐based ALK rearrangement assays in a similar cohort, the NGS‐based assay identified more ALK‐positive cases in NSCLC, suggesting that NGS was more sensitive than PCR for rearrangement detection. These data suggested that ALK rearrangement is more common in younger patients and that the relative frequency decreases along major age brackets, which is consistent with a previous report [31].
Based on different breakpoints in EML4 and ALK genes, EML4‐ALK can be divided into different subtypes [61], [62]. The most common subtypes were E13:A20 (1–13 exons in EML4 were fused with 20–29 exons in ALK, variant 1), E20:A20 (variant 2), and E6a/b:A20 (variant 3a/b), which were detected in 31%, 14.3%, and 36.9% of patients with EML4‐ALK NSCLC in the Chinese cohort, respectively. Clinical and preclinical studies have suggested differential clinical response to ALK inhibitors among different subtypes of EML4‐ALK. For example, EML4‐ALK variant 1 is associated with longer response duration to ALK inhibitors, whereas variant 3a/b is associated with earlier emergence of resistance to ALK inhibitors [61], [62]. Given that different variants respond differently to ALK inhibitors, the present findings may have important clinical implications. In the Chinese NSCLC population, the association between the incidence of ALK rearrangement and age was identified.
Conclusion
In the present study, genomic signature of lung cancer‐related driver genes identified by an NGS‐based target sequencing assay showed higher frequency of uncommon EGFR mutations than traditional Sanger sequencing or ARMS, and higher prevalence of ALK rearrangements but fewer KRAS mutations compared with those in the Western population. In total, 73.9% of Chinese patients with NSCLC harbored at least one genomic alteration in druggable genes recommended by the NCCN guideline. These findings revealed that more patients would have the opportunity to benefit from current targeted therapies approved by the FDA. NGS‐based targeted sequencing provided a more powerful tool to further fully understand the genomic features unique to Chinese patients.
See http://www.TheOncologist.com for supplemental material available online.
Acknowledgments
This work was supported by National Natural Science Foundation of China. (No. 81672415) and Natural Science Foundation of Guangdong Province, China. (No. 2017A030313474); The National Natural Science Foundation of China (No. 81871886); Grant [2013]163 from Key Laboratory of Malignant Tumor Molecular Mechanism and Translational Medicine of Guangzhou Bureau of Science and Information Technology; Grant KLB09001 from the Key Laboratory of Malignant Tumor Gene Regulation and Target Therapy of Guangdong Higher Education institutes; and a grant from Guangdong Science and Technology Department (2015B050501004).
Contributed equally.
Author Contributions
Conception/design: Kai Wang, Minghui Wang
Provision of study material or patients: Shiwang Wen, Lei Dai, Lei Wang, Wenjian Wang, Duoguang Wu, Kefeng Wang, Zhanghai He
Collection and/or assembly of data: Xiaowei Dong, Ming Yao
Data analysis and interpretation: Aodi Wang, Hui Chen, Peng Zhang
Manuscript writing: Yu‐An Dong, Kai Wang, Ming Yao
Final approval of manuscript: Shiwang Wen, Lei Dai, Lei Wang, Wenjian Wang, Duoguang Wu, Kefeng Wang, Zhanghai He, Aodi Wang, Hui Chen, Peng Zhang, Xiaowei Dong, Yu‐An Dong, Kai Wang, Ming Yao, Minghui Wang
Disclosures
Aodi Wang: OrigiMed (E); Hui Chen: OrigiMed (E); Peng Zhang: OrigiMed (E); Xiaowei Dong: OrigiMed (E); Yu‐An Dong: OrigiMed (E); Kai Wang: OrigiMed (E); Ming Yao: OrigiMed (E). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Torre LA, Bray F, Siegel RL et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017;67:7–30. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115–132. [DOI] [PubMed] [Google Scholar]
- 4.Morgensztern D, Campo MJ, Dahlberg SE et al. Molecularly targeted therapies in non‐small‐cell lung cancer annual update 2014. J Thorac Oncol 2015;10(1 suppl 1):S1–S63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han JY, Kim SH, Lee YS et al. Comparison of targeted next‐generation sequencing with conventional sequencing for predicting the responsiveness to epidermal growth factor receptor‐tyrosine kinase inhibitor (EGFR‐TKI) therapy in never‐smokers with lung adenocarcinoma. Lung Cancer 2014;85:161–167. [DOI] [PubMed] [Google Scholar]
- 6.Karnes HE, Duncavage EJ, Bernadt CT. Targeted next‐generation sequencing using fine‐needle aspirates from adenocarcinomas of the lung. Cancer Cytopathol 2014;122:104–113. [DOI] [PubMed] [Google Scholar]
- 7.Paez JG, Janne PA, Lee JC et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science 2004;304:1497–1500. [DOI] [PubMed] [Google Scholar]
- 8.Molina JR, Yang P, Cassivi SD et al. Non‐small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc 2008;83:584–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suh JH, Johnson A, Albacker L et al. Comprehensive genomic profiling facilitates implementation of the National Comprehensive Cancer Network guidelines for lung cancer biomarker testing and identifies patients who may benefit from enrollment in mechanism‐driven clinical trials. The Oncologist 2016;21:684–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Y, Au JS, Thongprasert S et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non‐small‐cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol 2014;9:154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gusterson BA, Hunter KD. Should we be surprised at the paucity of response to EGFR inhibitors? Lancet Oncol 2009;10:522–527. [DOI] [PubMed] [Google Scholar]
- 12.Douillard JY, Ostoros G, Cobo M et al. First‐line gefitinib in Caucasian EGFR mutation‐positive NSCLC patients: A phase‐IV, open‐label, single‐arm study. Br J Cancer 2014;110:55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dungo RT, Keating GM. Afatinib: First global approval. Drugs 2013;73:1503–1515. [DOI] [PubMed] [Google Scholar]
- 14.Shi Y, Zhang L, Liu X et al. Icotinib versus gefitinib in previously treated advanced non‐small‐cell lung cancer (ICOGEN): A randomised, double‐blind phase 3 non‐inferiority trial. Lancet Oncol 2013;14:953–961. [DOI] [PubMed] [Google Scholar]
- 15.Sequist LV, Yang JC, Yamamoto N et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013;31:3327–3334. [DOI] [PubMed] [Google Scholar]
- 16.Tsao MS, Sakurada A, Cutz JC et al. Erlotinib in lung cancer ‐ Molecular and clinical predictors of outcome. N Engl J Med 2005;353:133–144. [DOI] [PubMed] [Google Scholar]
- 17.Yu HA, Arcila ME, Rekhtman N et al. Analysis of tumor specimens at the time of acquired resistance to EGFR‐TKI therapy in 155 patients with EGFR‐mutant lung cancers. Clin Cancer Res 2013;19:2240–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sequist LV, Waltman BA, Dias‐Santagata D et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi S, Boggon TJ, Dayaram T et al. EGFR mutation and resistance of non‐small‐cell lung cancer to gefitinib. N Engl J Med 2005;352:786–792. [DOI] [PubMed] [Google Scholar]
- 20.Mok TS, Wu YL, Ahn MJ et al. Osimertinib or platinum‐pemetrexed in EGFR T790M‐positive lung cancer. N Engl J Med 2017;376:629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe S, Minegishi Y, Yoshizawa H et al. Effectiveness of gefitinib against non‐small‐cell lung cancer with the uncommon EGFR mutations G719X and l861Q. J Thorac Oncol 2014;9:189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiu CH, Yang CT, Shih JY et al. Epidermal growth factor receptor tyrosine kinase inhibitor treatment response in advanced lung adenocarcinomas with G719X/l861Q/S768I mutations. J Thorac Oncol 2015;10:793–799. [DOI] [PubMed] [Google Scholar]
- 23.Yang JC, Sequist LV, Geater SL et al. Clinical activity of afatinib in patients with advanced non‐small‐cell lung cancer harbouring uncommon EGFR mutations: A combined post‐hoc analysis of LUX‐Lung 2, LUX‐Lung 3, and LUX‐Lung 6. Lancet Oncol 2015;16:830–838. [DOI] [PubMed] [Google Scholar]
- 24.Muller IB, de Langen AJ, Giovannetti E et al. Anaplastic lymphoma kinase inhibition in metastatic non‐small cell lung cancer: Clinical impact of alectinib. Onco Targets Ther 2017;10:4535–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nayak AK, Kumar V, Ma T et al. Magnetic antiskyrmions above room temperature in tetragonal heusler materials. Nature 2017;548:561–566. [DOI] [PubMed] [Google Scholar]
- 26.Bergethon K, Shaw AT, Ou SH et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol 2012;30:863–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lipson D, Capelletti M, Yelensky R et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med 2012;18:382–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davies KD, Le AT, Theodoro MF et al. Identifying and targeting ROS1 gene fusions in non‐small cell lung cancer. Clin Cancer Res 2012;18:4570–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeuchi K, Soda M, Togashi Y et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med 2012;18:378–381. [DOI] [PubMed] [Google Scholar]
- 30.Kwak EL, Bang YJ, Camidge DR et al. Anaplastic lymphoma kinase inhibition in non‐small‐cell lung cancer. N Engl J Med 2010;363:1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaw AT, Yeap BY, Mino‐Kenudson M et al. Clinical features and outcome of patients with non‐small‐cell lung cancer who harbor EML4‐ALK. J Clin Oncol 2009;27:4247–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim HR, Lim SM, Kim HJ et al. The frequency and impact of ROS1 rearrangement on clinical outcomes in never smokers with lung adenocarcinoma. Ann Oncol 2013;24:2364–2370. [DOI] [PubMed] [Google Scholar]
- 33.Mazieres J, Zalcman G, Crino L et al. Crizotinib therapy for advanced lung adenocarcinoma and a ROS1 rearrangement: Results from the EUROS1 cohort. J Clin Oncol 2015;33:992–999. [DOI] [PubMed] [Google Scholar]
- 34.Solomon BJ, Mok T, Kim DW et al. First‐line crizotinib versus chemotherapy in ALK‐positive lung cancer. N Engl J Med 2014;371:2167–2177. [DOI] [PubMed] [Google Scholar]
- 35.Kazandjian D, Blumenthal GM, Chen HY et al. FDA approval summary: Crizotinib for the treatment of metastatic non‐small cell lung cancer with anaplastic lymphoma kinase rearrangements. The Oncologist 2014;19:e5–e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaw AT, Kim DW, Nakagawa K et al. Crizotinib versus chemotherapy in advanced ALK‐positive lung cancer. N Engl J Med 2013;368:2385–2394. [DOI] [PubMed] [Google Scholar]
- 37.Drilon A, Rekhtman N, Arcila M et al. Cabozantinib in patients with advanced RET‐rearranged non‐small‐cell lung cancer: An open‐label, single‐centre, phase 2, single‐arm trial. Lancet Oncol 2016;17:1653–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee SH, Lee JK, Ahn MJ et al. Vandetanib in pretreated patients with advanced non‐small cell lung cancer‐harboring RET rearrangement: A phase II clinical trial. Ann Oncol 2017;28:292–297. [DOI] [PubMed] [Google Scholar]
- 39.Drilon A, Wang L, Hasanovic A et al. Response to cabozantinib in patients with RET fusion‐positive lung adenocarcinomas. Cancer Discov 2013;3:630–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tu HY, Ke EE, Yang JJ et al. A comprehensive review of uncommon EGFR mutations in patients with non‐small cell lung cancer. Lung Cancer 2017;114:96–102. [DOI] [PubMed] [Google Scholar]
- 41.Chen K, Yu X, Wang H et al. Uncommon mutation types of epidermal growth factor receptor and response to EGFR tyrosine kinase inhibitors in Chinese non‐small cell lung cancer patients. Cancer Chemother Pharmacol 2017;80:1179–1187. [DOI] [PubMed] [Google Scholar]
- 42.Campbell JD, Alexandrov A, Kim J et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48:607–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho J, Bass AJ, Lawrence MS et al. Colon cancer‐derived oncogenic EGFR G724S mutant identified by whole genome sequence analysis is dependent on asymmetric dimerization and sensitive to cetuximab. Mol Cancer 2014;13:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oztan A, Fischer S, Schrock AB et al. Emergence of EGFR G724S mutation in EGFR‐mutant lung adenocarcinoma post progression on osimertinib. Lung Cancer 2017;111:84–87. [DOI] [PubMed] [Google Scholar]
- 45.Lou Y, Pecot CV, Tran HT et al. Germline mutation of T790M and dual/multiple EGFR mutations in patients with lung adenocarcinoma. Clin Lung Cancer 2016;17:e5–e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu HA, Arcila ME, Harlan Fleischut M et al. Germline EGFR T790M mutation found in multiple members of a familial cohort. J Thorac Oncol 2014;9:554–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gazdar A, Robinson L, Oliver D et al. Hereditary lung cancer syndrome targets never smokers with germline EGFR gene T790M mutations. J Thorac Oncol 2014;9:456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu SY, Mok T, Wu YL. Novel targeted agents for the treatment of lung cancer in China. Cancer 2015;121(suppl 17):3089–3096. [DOI] [PubMed] [Google Scholar]
- 49.Kris MG, Johnson BE, Berry LD et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014;311:1998–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu YL, Zhou Q. Clinical trials and biomarker research on lung cancer in China. Expert Opin Ther Targets 2012;16(suppl 1):S45–S50. [DOI] [PubMed] [Google Scholar]
- 51.Suh JH, Schrock AB, Johnson A et al. Hybrid capture‐based comprehensive genomic profiling identifies lung cancer patients with well‐characterized sensitizing epidermal growth factor receptor point mutations that were not detected by standard of care testing. The Oncologist 2018;23:776–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oxnard GR, Lo PC, Nishino M et al. Natural history and molecular characteristics of lung cancers harboring EGFR exon 20 insertions. J Thorac Oncol 2013;8:179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lund‐Iversen M, Kleinberg L, Fjellbirkeland L et al. Clinicopathological characteristics of 11 NSCLC patients with EGFR‐exon 20 mutations. J Thorac Oncol 2012;7:1471–1473. [DOI] [PubMed] [Google Scholar]
- 54.Huang L, Fu L. Mechanisms of resistance to EGFR tyrosine kinase inhibitors. Acta Pharm Sin B 2015;5:390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, Hu H, Wang R et al. Primary concomitant EGFR T790M mutation predicted worse prognosis in non‐small cell lung cancer patients. Onco Targets Ther 2014;7:513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi J, Yang H, Jiang T et al. Uncommon EGFR mutations in a cohort of chinese NSCLC patients and outcomes of first‐line EGFR‐TKIs and platinum‐based chemotherapy. Chin J Cancer Res 2017;29:543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hou H, Zhu H, Zhao H et al. Comprehensive molecular characterization of young Chinese patients with lung adenocarcinoma identified a distinctive genetic profile. The Oncologist 2018;23:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kobayashi S, Canepa HM, Bailey AS et al. Compound EGFR mutations and response to EGFR tyrosine kinase inhibitors. J Thorac Oncol 2013;8:45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Z, Cheng Y, An T et al. Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first‐line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): A phase 2, single‐arm, multicentre clinical trial. Lancet Respir Med 2018;6:681–690. [DOI] [PubMed] [Google Scholar]
- 60.Ross JS, Ali SM, Fasan O et al. ALK fusions in a wide variety of tumor types respond to anti‐ALK targeted therapy. The Oncologist 2017;22:1444–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woo CG, Seo S, Kim SW et al. Differential protein stability and clinical responses of EML4‐ALK fusion variants to various ALK inhibitors in advanced ALK‐rearranged non‐small cell lung cancer. Ann Oncol 2017;28:791–797. [DOI] [PubMed] [Google Scholar]
- 62.S‐HI Ou, Schrock AB, Gowen K et al. Association of ALK resistance mutations by EML4‐ALK variant (v3 vs. Non‐v3) in ALK+ non‐small cell lung cancer (NSCLC). J Clin Oncol 2017;35(suppl 15):9010a. [Google Scholar]