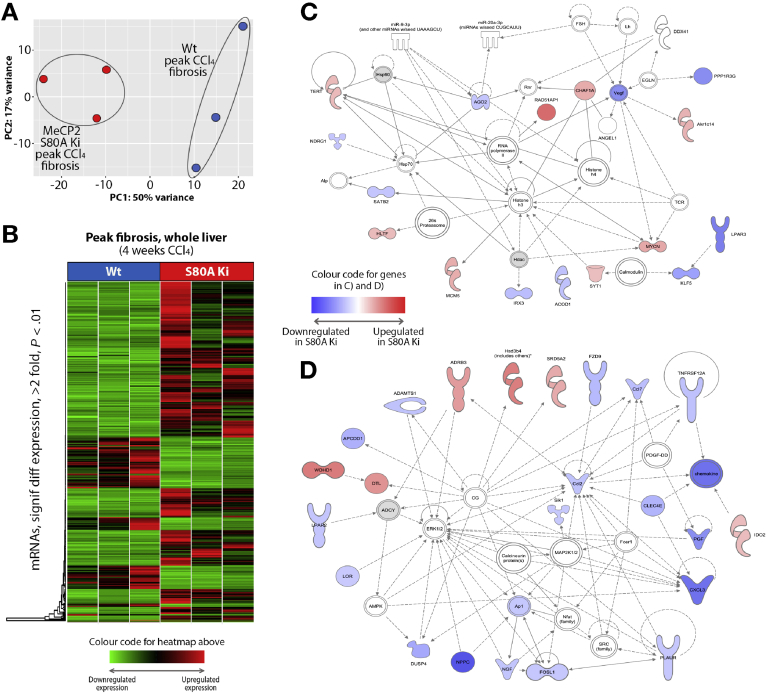
Figure 7.
Mecp2S80 phosphorylation regulates gene expression in CCl4-induced liver fibrosis. (A) Gene expression variances between WT and Mecp2S80A fibrotic livers displayed as PCA plots of scaled log2-transformed transcript counts. (B) Heatmap displaying results of transcriptome sequencing carried out using 3 CCl4-injured livers from each of the WT and Mecp2S80A mice; genes with greater than 2-fold change and significant to P < .01 are shown. Green indicates negative values (down-regulated in Mecp2S80A); red, positive (up-regulated in Mecp2S80A); black, unchanged. (C, D) IPA was used to form a network of focused genes that are downstream targets of differentially expressed RNAs. Blue nodes show down-regulated genes in Mecp2S80A CCl4-injured liver; red nodes are up-regulated. Symbol shapes signify the nature of the encoded protein (see Supplementary Figures 1 and 2 for shape information), and unshaded symbols signify genes relevant to the pathway but not differentially expressed in our data set. signif diff, significantly different.