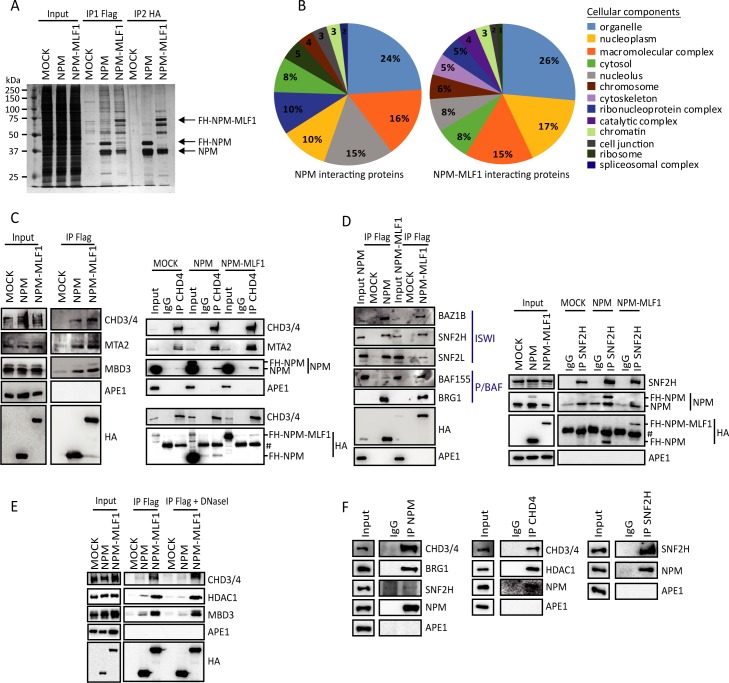
Fig 1. NPM and NPM-MLF1 interact with chromatin remodeling complexes.
A) Flag-HA tandem immunoaffinity purification; nuclear extract from MOCK, NPM or NPM-MLF1 expressing cells were used for sequential immunoaffinity purification and a fraction of the different purification steps was loaded and detected by silver staining; Input: nuclear extracts; IP1 Flag: Flag purification; IP2 HA: HA purification; arrows indicate the bands corresponding to FH-NPM-MLF1, FH-NPM and the endogenous NPM; protein molecular weights are indicated on the left side of the panel; B) LC-MS/MS data analysis made with STRING database; NPM and NPM-MLF1 interacting partners are classified according to cellular component organization; C, D) Co-immunoprecipitation assays; K562 nuclear extracts of MOCK, NPM or NPM-MLF1 expressing cells were immunoprecipitated (IP) with Flag, CHD4, SNF2H antibodies or isotype-matched immunoglobulins (IgG); the antibodies used for Western blots are indicated on the right side of the panels; antibodies were specific for NuRD (panel C) or ISWI and P/BAF (panel D) complex; APE1 was used as negative control; E) Immunoprecipitation assay with Flag antibodies; K562 nuclear extracts of MOCK, NPM or NPM-MLF1 were treated with 10 μg/mL of DNaseI; F) Co-immunoprecipitation experiments were performed with NPM, BRG1, SNF2H antibodies or isotype-matched immunoglobulin (IgG) in HEL WT cell nuclear extracts. The proteins detected by Western blot are indicated on the right side of the panels.