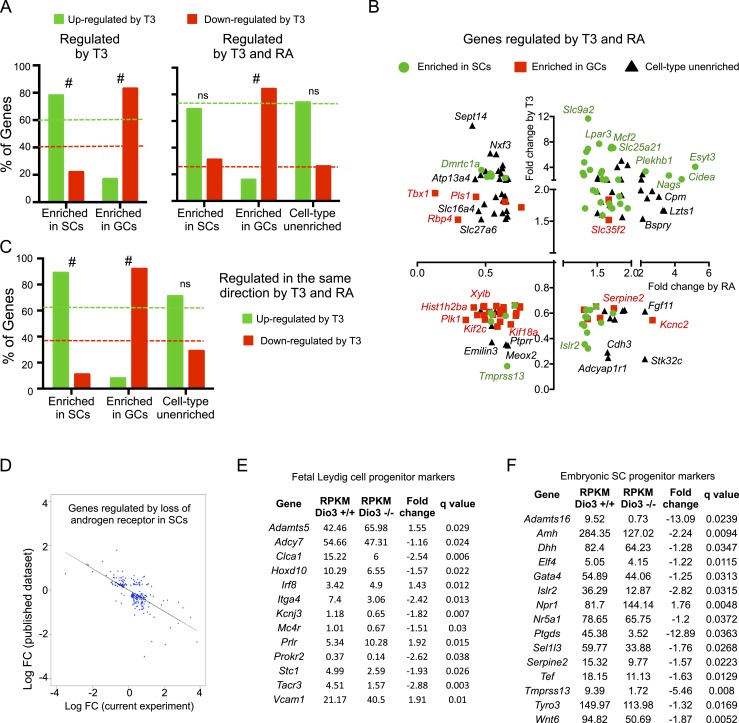
Figure 5.
DEGs in Dio3−/− testis that were also regulated in other testis gene profiling data sets. (A–C) Comparison with data reported by Evans et al. (48). (A) Percentage distribution of T3 upregulation and downregulation of genes according to cell type enrichment for genes regulated by T3 (left) or regulated by both T3 and RA (right). (B) T3-dependent vs RA-dependent fold change in gene expression for 155 genes regulated by T3 and RA, depicted according to cell type enrichment. The names of some of the genes are noted. (C) Percentage distribution of T3 upregulation and downregulation of genes according to cell type enrichment for 91 genes that are regulated in the same direction by both T3 and RA. Dotted lines indicate the percentage of upregulated and downregulated genes (green and red, respectively) in the full set of relevant genes. #P < 0.001, as determined by a χ2 test. (D) DEGs also regulated in a mouse model of androgen receptor deficiency in SCs (50). The expression changes in each model show a strong inverse correlation. (E and F) Data on DEGs also identified by transcriptome analyses (51) as markers of progenitor cells for fetal (E) LCs and (F) SCs. The vast majority of those markers were downregulated in the current study. ns, not significant. [From: Green CD, Ma Q, Manske GL, Shami AN, Zheng X, Marini S, Moritz L, Sultan C, Gurczynski SJ, Moore BB, Tallquist MD, Li JZ, Hammoud SS. A comprehensive roadmap of murine spermatogenesis defined by single-cell RNA-seq. Developmental Cell 2018;46:651–667.e610.]