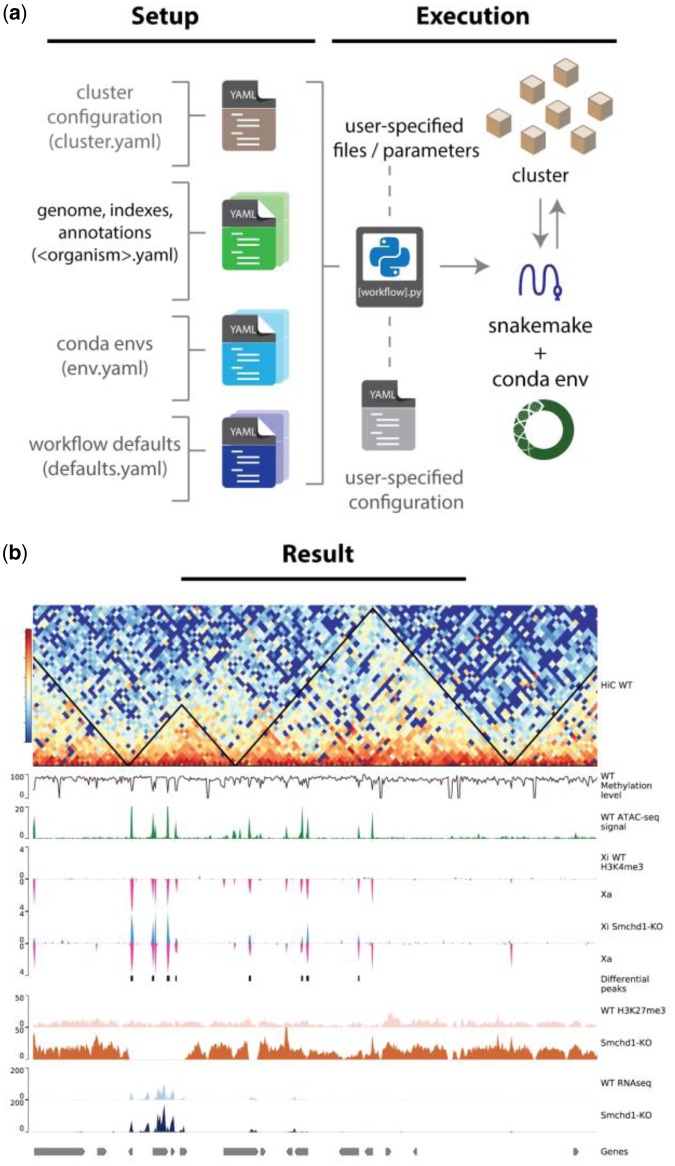
Fig. 1.
Setup, execution and results from snakePipes. (a) All configurable parameters for snakepipes are defined as YAML files during setup. However, most parameters can be overwritten during execution by providing another YAML file, adding flexibility to the analysis. (b) Output of HiC (track 1), WGBS (track 2), ATAC-seq (track 3), allele-specific ChIP-seq (tracks 3–7) and RNA-seq (tracks 8–9) workflows, plotted using pyGenomeTracks (Ramírez et al., 2018)