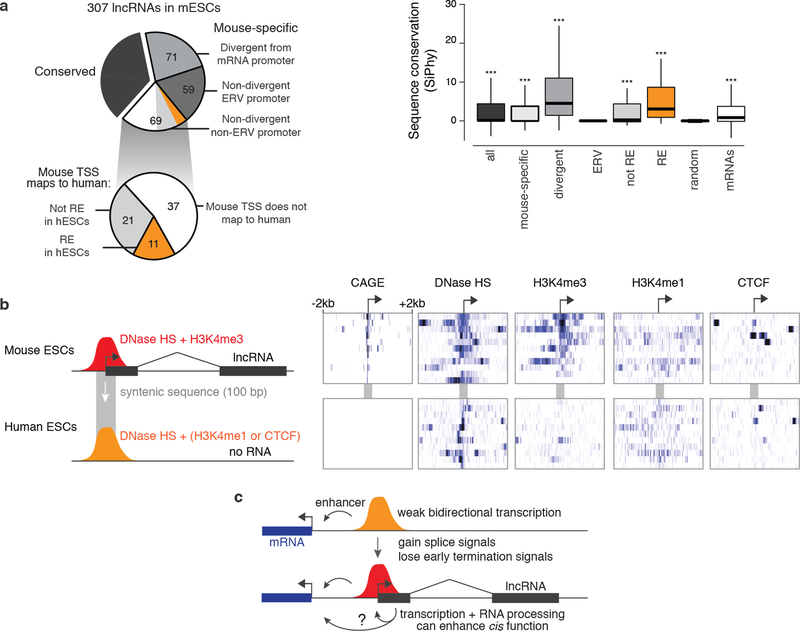
Extended Data Fig. 10. Classification of lncRNAs based on conservation and promoter location.
(a) Classification of 307 lncRNAs expressed in mESCs. “Conserved” transcripts are those that show significant evidence of capped analysis of gene expression (CAGE) data and/or p(A)+ RNA in syntenic loci (see Methods). Divergent: initiating within 500 bp of an mRNA TSS, on the opposite strand. ERV: endogenous retroviral repetitive element (see Note S9). Boxplot shows sequence-level conservation of the promoters of subsets of lncRNAs expressed in mESCs. Random intergenic regions are matched to lncRNA promoters by GC content. Positive SiPhy score indicates evolutionary constraint on functional sequences. Orange category corresponds to mouse-specific lncRNAs that appear to have evolved from ancestral regulatory elements (REs) and correspond to sequences that show evidence for DNase I hypersensitivity in human embryonic stem cells. Significance is calculated compared to random intergenic regions using a Mann-Whitney U-test. ***: P < 0.001. Whiskers represent data within 1.5× the interquartile range of the box. (b) Chromatin and RNA data for 11 mouse-specific lncRNAs that appear to have evolved from ancestral regulatory elements. In mouse, these elements show evidence for CAGE, H3K4me3, and DNase I hypersensitivity, consistent with their roles as promoters. The syntenic sequences in human do not show evidence for CAGE but nonetheless are DNase I hypersensitive and are frequently marked by H3K4me1 and/or CTCF. (c) Model for evolution of lncRNAs from pre-existing enhancers, which often initiate weak bidirectional transcription to produce eRNA. Spliced transcripts may neutrally appear through the appearance of splice signals and loss of polyadenylation signals. In some cases, transcription, splicing, or other RNA processing mechanisms may feed back and contribute to the cis regulatory function of the promoter, producing a lncRNA as a byproduct.