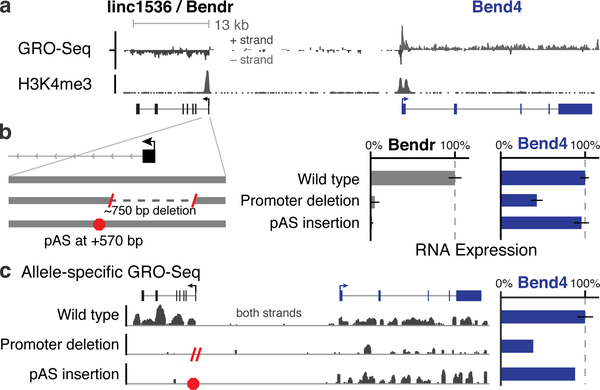
Fig. 2. Enhancer-like function of the Bendr promoter.
(a) Transcriptionally engaged RNA polymerase (GRO-Seq) and H3K4me3 occupancy (ChIP-Seq). (b) p(A)+ RNA expression upon deleting the Bendr promoter or inserting a pAS on modified versus unmodified alleles. Error bars: 95% CI for the mean (n ≥ 2 alleles, see Table S1). (c) Allele-specific GRO-seq signal for clones carrying the indicated modifications. Both clones are modified on the 129 allele, and only reads specifically mapping that allele are shown. Y-axis: normalized read count. Bar plot quantifies signal at Bend4, including 7 additional wild-type controls not shown on left.